In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA. The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences, and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome.

A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size.
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functional regions of the non-coding DNA fraction include regulatory sequences that control gene expression; scaffold attachment regions; origins of DNA replication; centromeres; and telomeres. Some non-coding regions appear to be mostly nonfunctional, such as introns, pseudogenes, intergenic DNA, and fragments of transposons and viruses. Regions that are completely nonfunctional are called junk DNA.

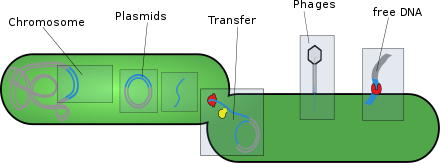
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher-order evolution while more significantly shifting perspectives on bacterial evolution.
Repeated sequences are short or long patterns that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres.

Retrotransposons are mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged.
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.
Exon shuffling is a molecular mechanism for the formation of new genes. It is a process through which two or more exons from different genes can be brought together ectopically, or the same exon can be duplicated, to create a new exon-intron structure. There are different mechanisms through which exon shuffling occurs: transposon mediated exon shuffling, crossover during sexual recombination of parental genomes and illegitimate recombination.
In molecular biology, insertional mutagenesis is the creation of mutations in DNA by the addition of one or more base pairs. Such insertional mutations can occur naturally, mediated by viruses or transposons, or can be artificially created for research purposes in the lab.

The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome.
Transposon mutagenesis, or transposition mutagenesis, is a biological process that allows genes to be transferred to a host organism's chromosome, interrupting or modifying the function of an extant gene on the chromosome and causing mutation. Transposon mutagenesis is much more effective than chemical mutagenesis, with a higher mutation frequency and a lower chance of killing the organism. Other advantages include being able to induce single hit mutations, being able to incorporate selectable markers in strain construction, and being able to recover genes after mutagenesis. Disadvantages include the low frequency of transposition in living systems, and the inaccuracy of most transposition systems.
Helitrons are one of the three groups of eukaryotic class 2 transposable elements (TEs) so far described. They are the eukaryotic rolling-circle transposable elements which are hypothesized to transpose by a rolling circle replication mechanism via a single-stranded DNA intermediate. They were first discovered in plants and in the nematode Caenorhabditis elegans, and now they have been identified in a diverse range of species, from protists to mammals. Helitrons make up a substantial fraction of many genomes where non-autonomous elements frequently outnumber the putative autonomous partner. Helitrons seem to have a major role in the evolution of host genomes. They frequently capture diverse host genes, some of which can evolve into novel host genes or become essential for Helitron transposition.
Transposons are semi-parasitic DNA sequences which can replicate and spread through the host's genome. They can be harnessed as a genetic tool for analysis of gene and protein function. The use of transposons is well-developed in Drosophila and in Thale cress and bacteria such as Escherichia coli.
The Sleeping Beauty transposon system is a synthetic DNA transposon designed to introduce precisely defined DNA sequences into the chromosomes of vertebrate animals for the purposes of introducing new traits and to discover new genes and their functions. It is a Tc1/mariner-type system, with the transposase resurrected from multiple inactive fish sequences.
The PiggyBac (PB) transposon system employs a genetically engineered transposase enzyme to insert a gene into a cell's genome. It is built upon the natural PiggyBac (PB) transposable element (transposon), enabling the back and forth movement of genes between chromosomes and genetic vectors such as plasmids through a "cut and paste" mechanism. During transposition, the PB transposase recognizes transposon-specific inverted terminal repeat sequences (ITRs) located on both ends of the transposon vector and efficiently moves the contents from the original sites and integrates them into TTAA chromosomal sites. The powerful activity of the PiggyBac transposon system enables genes of interest between the two ITRs in the PB vector to be easily mobilized into target genomes. The TTAA-specific transposon piggyBac is rapidly becoming a highly useful transposon for genetic engineering of a wide variety of species, particularly insects. They were discovered in 1989 by Malcolm Fraser at the University of Notre Dame.

Transposition is the process by which a specific genetic sequence, known as a transposon, is moved from one location of the genome to another. Simple, or conservative transposition, is a non-replicative mode of transposition. That is, in conservative transposition the transposon is completely removed from the genome and reintegrated into a new, non-homologous locus, the same genetic sequence is conserved throughout the entire process. The site in which the transposon is reintegrated into the genome is called the target site. A target site can be in the same chromosome as the transposon or within a different chromosome. Conservative transposition uses the "cut-and-paste" mechanism driven by the catalytic activity of the enzyme transposase. Transposase acts like DNA scissors; it is an enzyme that cuts through double-stranded DNA to remove the transposon, then transfers and pastes it into a target site.
DNA transposons are DNA sequences, sometimes referred to "jumping genes", that can move and integrate to different locations within the genome. They are class II transposable elements (TEs) that move through a DNA intermediate, as opposed to class I TEs, retrotransposons, that move through an RNA intermediate. DNA transposons can move in the DNA of an organism via a single-or double-stranded DNA intermediate. DNA transposons have been found in both prokaryotic and eukaryotic organisms. They can make up a significant portion of an organism's genome, particularly in eukaryotes. In prokaryotes, TE's can facilitate the horizontal transfer of antibiotic resistance or other genes associated with virulence. After replicating and propagating in a host, all transposon copies become inactivated and are lost unless the transposon passes to a genome by starting a new life cycle with horizontal transfer. DNA transposons do not randomly insert themselves into the genome, but rather show preference for specific sites.
Polintons are large DNA transposons which contain genes with homology to viral proteins and which are often found in eukaryotic genomes. They were first discovered in the mid-2000s and are the largest and most complex known DNA transposons. Polintons encode up to 10 individual proteins and derive their name from two key proteins, a DNA polymerase and a retroviral-like integrase.
Integrative and conjugative elements (ICEs) are mobile genetic elements present in both gram-positive and gram-negative bacteria. In a donor cell, ICEs are located primarily on the chromosome, but have the ability to excise themselves from the genome and transfer to recipient cells via bacterial conjugation.
Retrozymes are a family of retrotransposons first discovered in the genomes of plants but now also known in genomes of animals. Retrozymes contain a hammerhead ribozyme (HHR) in their sequences, although they do not possess any coding regions. Retrozymes are nonautonomous retroelements, and so borrow proteins from other elements to move into new regions of a genome. Retrozymes are actively transcribed into covalently closed circular RNAs and are detected in both polarities, which may indicate the use of rolling circle replication in their lifecycle.