Antimycins are produced as secondary metabolites by Streptomyces bacteria, a soil bacteria. These specialized metabolites likely function to kill neighboring organisms in order to provide the streptomyces bacteria with a competitive edge.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.
Biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined together to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
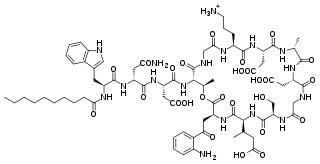
Daptomycin is a lipopeptide antibiotic used in the treatment of systemic and life-threatening infections caused by Gram-positive organisms. It is a naturally occurring compound found in the soil saprotroph Streptomyces roseosporus. Its distinct mechanism of action makes it useful in treating infections caused by multiple drug-resistant bacteria. It is marketed in the United States under the trade name Cubicin by Cubist Pharmaceuticals.
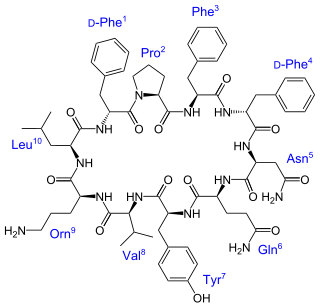
Tyrocidine is a mixture of cyclic decapeptides produced by the bacteria Bacillus brevis found in soil. It can be composed of 4 different amino acid sequences, giving tyrocidine A–D. Tyrocidine is the major constituent of tyrothricin, which also contains gramicidin. Tyrocidine was the first commercially available antibiotic, but has been found to be toxic toward human blood and reproductive cells. The function of tyrocidine within its host B. brevis is thought to be regulation of sporulation.

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can only synthesize 11 of the 20 standard amino acids, and in time of accelerated growth, histidine, can be considered an essential amino acid.

Dihydropteroate synthase is an enzyme classified under EC 2.5.1.15. It produces dihydropteroate in bacteria, but it is not expressed in most eukaryotes including humans. This makes it a useful target for sulfonamide antibiotics, which compete with the PABA precursor.

Enterobactin is a high affinity siderophore that acquires iron for microbial systems. It is primarily found in Gram-negative bacteria, such as Escherichia coli and Salmonella typhimurium.

The Tryptophan operon leader is an RNA element found at the 5′ of some bacterial tryptophan operons. The leader sequence can assume two different secondary structures known as the terminator and the anti-terminator structure. The leader also codes for very short peptide sequence that is rich in tryptophan. The terminator structure is recognised as a termination signal for RNA polymerase and the operon is not transcribed. This structure forms when the cell has an excess of tryptophan and ribosome movement over the leader transcript is not impeded. When there is a deficiency of the charged tryptophanyl tRNA the ribosome translating the leader peptide stalls and the antiterminator structure can form. This allows RNA polymerase to transcribe the operon.
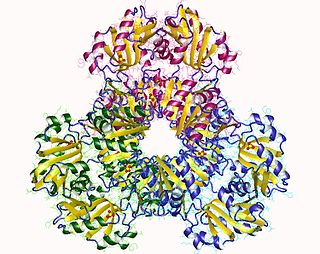
Ribose-phosphate diphosphokinase is an enzyme that converts ribose 5-phosphate into phosphoribosyl pyrophosphate (PRPP). It is classified under EC 2.7.6.1.
In enzymology, a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) is an enzyme that catalyzes the chemical reaction
In enzymology, a (2,3-dihydroxybenzoyl)adenylate synthase is an enzyme that catalyzes the chemical reaction

In enzymology, a riboflavin kinase is an enzyme that catalyzes the chemical reaction
HutP is one of the anti-terminator proteins of Bacillus subtilis, which is responsible for regulating the expression of the hut structural genes of this organism in response to changes in the intracellular levels of L-histidine and divalent metal ions. In the hut operon, HutP is located just downstream from the promoter, while the five other subsequent structural genes, hutH, hutU, hutI, hutG and hutM, are positioned far downstream from the promoter. In the presence of L-histidine and divalent metal ions, HutP binds to the nascent hut mRNA leader transcript. This allows the anti-terminator to form, thereby preventing the formation of the terminator and permitting transcriptional read-through into the hut structural genes. In the absence of L-histidine and divalent metal ions or both, HutP does not bind to the hut mRNA, thus allowing the formation of a stem loop terminator structure within the nucleotide sequence located between the hutP and structural genes.

Bacillus anthracis is the agent of anthrax—a common disease of livestock and, occasionally, of Humans—and the only obligate pathogen within the genus Bacillus. This disease can be classified as a zoonosis, causing infected animals to transmit the disease to humans. B. anthracis is a Gram-positive, endospore-forming, rod-shaped bacterium, with a width of 1.0–1.2 µm and a length of 3–5 µm. It can be grown in an ordinary nutrient medium under aerobic or anaerobic conditions.
Streptogramin B is a subgroup of the streptogramin antibiotics family. These natural products are cyclic hexa- or hepta depsipeptides produced by various members of the genus of bacteria Streptomyces. Many of the members of the streptogramins reported in the literature have the same structure and different names; for example, pristinamycin IA = vernamycin Bα = mikamycin B = osteogrycin B.

In molecular biology, the Cofactor transferase family is a family of protein domains that includes biotin protein ligases, lipoate-protein ligases A, octanoyl-(acyl carrier protein):protein N-octanoyltransferases, and lipoyl-protein:protein N-lipoyltransferases. The metabolism of the cofactors Biotin and lipoic acid share this family. They also share the target modification domain, and the sulfur insertion enzyme.
Many bacteria secrete small iron-binding molecules called siderophores, which bind strongly to ferric ions. FepA is an integral bacterial outer membrane porin protein that belongs to Outer membrane receptor family and provides the active transport of iron bound by the siderophore enterobactin from the extracellular space, into the periplasm of Gram-negative bacteria. FepA has also been shown to transport vitamin B12, and colicins B and D as well. This protein belongs to family of ligand-gated protein channels.
Ribosomally synthesized and post-translationally modified peptides (RiPPs), also known as ribosomal natural products, are a diverse class of natural products of ribosomal origin. Consisting of more than 20 sub-classes, RiPPs are produced by a variety of organisms, including prokaryotes, eukaryotes, and archaea, and they possess a wide range of biological functions.

Vibriobactin is a catechol siderophore that helps the microbial system to acquire iron. It was first isolated from Vibrio cholerae.












