| |
| Identifiers | |
|---|---|
| |
| CAS Number | |
| PubChem CID | |
| ChemSpider | |
| UNII | |
| KEGG | |
| ChEBI | |
| ChEMBL | |
| CompTox Dashboard (EPA) | |
| Chemical and physical data | |
| Formula | C23H35NOS |
| Molar mass | 373.60 g·mol−1 |
| 3D model (JSmol) | |
| |
| |
| (verify) | |
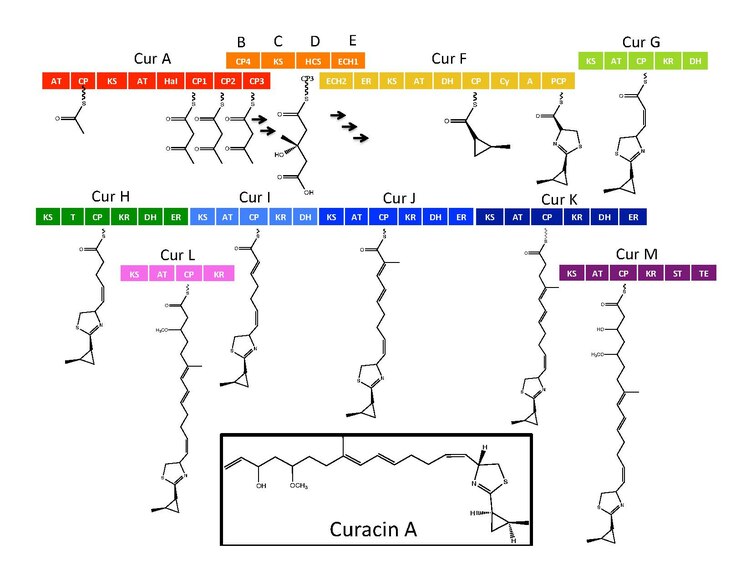
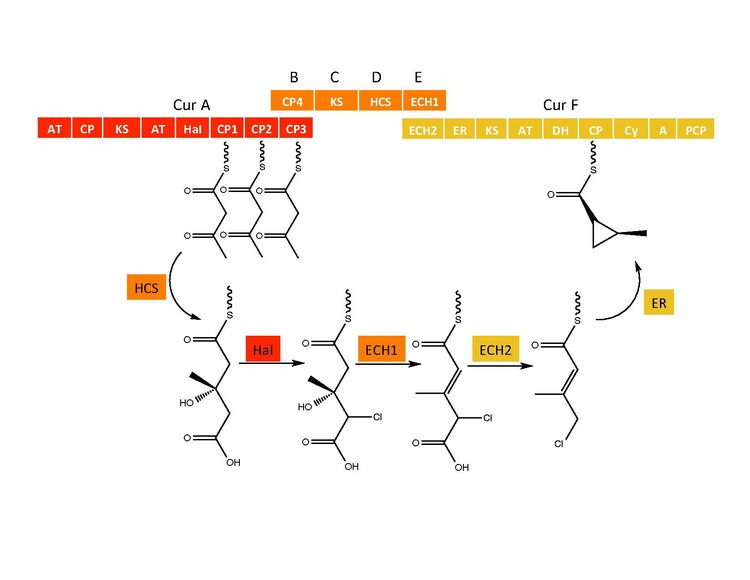
Curacin A is a hybrid polyketide synthase (PKS)/nonribosomal peptide synthase (NRPS) derived natural product produced isolated from the cyanobacterium Lyngbya majuscula . [1] Curacin A belongs to a family of natural products including jamaicamide, mupirocin, and pederin that have an unusual terminal alkene. Additionally, Curacin A contains a notable thiazoline ring and a unique cyclopropyl moiety, which is essential to the compound's biological activity. [1] [2] Curacin A has been characterized as potent antiproliferative cytotoxic compound with notable anticancer activity for several cancer lines including renal, colon, and breast cancer. [2] [3] Curacin A has been shown to interact with colchicine binding sites on tubulin, which inhibits microtubule polymerization, an essential process for cell division and proliferation. [1] [4]