
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, USA.

Fas ligand is a type-II transmembrane protein expressed on various types of cells, including cytotoxic T lymphocytes, monocytes, neutrophils, breast epithelial cells, vascular endothelial cells and natural killer (NK) cells. It binds with its receptor, called FAS receptor and plays a crucial role in the regulation of the immune system and in induction of apoptosis, a programmed cell death.

The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways. The DED domain is found in inactive procaspases and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis. Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the Death domain (DD).

In the field of cell biology, TNF-related apoptosis-inducing ligand (TRAIL), is a protein functioning as a ligand that induces the process of cell death called apoptosis.

Amos Bairoch is a Swiss bioinformatician and Professor of Bioinformatics at the Department of Human Protein Sciences of the University of Geneva where he leads the CALIPHO group at the Swiss Institute of Bioinformatics (SIB) combining bioinformatics, curation, and experimental efforts to functionally characterize human proteins.
InterPro is a database of protein families, protein domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.

Caspase recruitment domains, or caspase activation and recruitment domains (CARDs), are interaction motifs found in a wide array of proteins, typically those involved in processes relating to inflammation and apoptosis. These domains mediate the formation of larger protein complexes via direct interactions between individual CARDs. CARDs are found on a strikingly wide range of proteins, including helicases, kinases, mitochondrial proteins, caspases, and other cytoplasmic factors.

The death fold is a tertiary structure motif commonly found in proteins involved in apoptosis or inflammation-related processes. This motif is commonly found in domains that participate in protein–protein interactions leading to the formation of large functional complexes. Examples of death fold domains include the death domain (DD), death effector domain (DED), caspase recruitment domain (CARD), and pyrin domain (PYD).
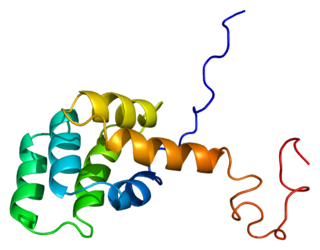
The Fas receptor, also known as Fas, FasR, apoptosis antigen 1, cluster of differentiation 95 (CD95) or tumor necrosis factor receptor superfamily member 6 (TNFRSF6), is a protein that in humans is encoded by the FAS gene. Fas was first identified using a monoclonal antibody generated by immunizing mice with the FS-7 cell line. Thus, the name Fas is derived from FS-7-associated surface antigen.

FAS-associated death domain protein, also called MORT1, is encoded by the FADD gene on the 11q13.3 region of chromosome 11 in humans.

In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.
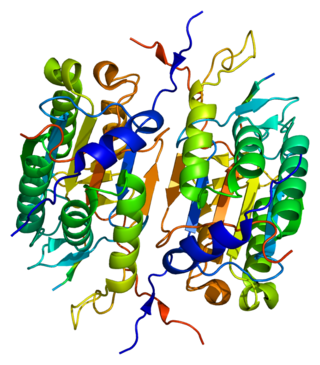
Caspase 2 also known as CASP2 is an enzyme that, in humans, is encoded by the CASP2 gene. CASP2 orthologs have been identified in nearly all mammals for which complete genome data are available. Unique orthologs are also present in birds, lizards, lissamphibians, and teleosts.

MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels. MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the Lawrence Berkeley National Laboratory in Berkeley, California. The site was launched in 2005, with regular updates until 2011.
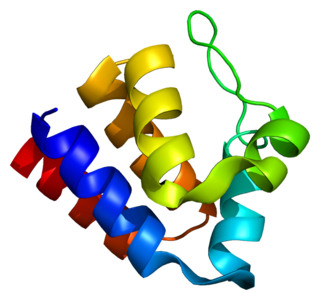
PYCARD, often referred to as ASC, is a protein that in humans is encoded by the PYCARD gene. It is localized mainly in the nucleus of monocytes and macrophages. In case of pathogen infection, however, it relocalizes rapidly to the cytoplasm, perinuclear space, endoplasmic reticulum and mitochondria and it is a key adaptor protein in activation of the inflammasome.
SUPERFAMILY is a database and search platform of structural and functional annotation for all proteins and genomes. It classifies amino acid sequences into known structural domains, especially into SCOP superfamilies. Domains are functional, structural, and evolutionary units that form proteins. Domains of common Ancestry are grouped into superfamilies. The domains and domain superfamilies are defined and described in SCOP. Superfamilies are groups of proteins which have structural evidence to support a common evolutionary ancestor but may not have detectable sequence homology.

The death domain (DD) is a protein interaction module composed of a bundle of six alpha-helices. DD is a subclass of protein motif known as the death fold and is related in sequence and structure to the death effector domain (DED) and the caspase recruitment domain (CARD), which work in similar pathways and show similar interaction properties. DD bind each other forming oligomers. Mammals have numerous and diverse DD-containing proteins. Within these proteins, the DD domains can be found in combination with other domains, including: CARDs, DEDs, ankyrin repeats, caspase-like folds, kinase domains, leucine zippers, leucine-rich repeats (LRR), TIR domains, and ZU5 domains.

A pyrin domain is a protein domain and a subclass of protein motif known as the death fold, the 4th and most recently discovered member of the death domain superfamily (DDF). It was originally discovered in the pyrin protein, or marenostrin, encoded by MEFV. The mutation of the MEFV gene is the cause of the disease known as Familial Mediterranean Fever. The domain is encoded in 23 human proteins and at least 31 mouse genes.
PDBsum is a database that provides an overview of the contents of each 3D macromolecular structure deposited in the Protein Data Bank (PDB).
TopFIND is the Termini oriented protein Function Inferred Database (TopFIND) is an integrated knowledgebase focused on protein termini, their formation by proteases and functional implications. It contains information about the processing and the processing state of proteins and functional implications thereof derived from research literature, contributions by the scientific community and biological databases.
In bioinformatics, the PANTHER classification system is a large curated biological database of gene/protein families and their functionally related subfamilies that can be used to classify and identify the function of gene products. PANTHER is part of the Gene Ontology Reference Genome Project designed to classify proteins and their genes for high-throughput analysis.














