
Escherichia coli, also known as E. coli, is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes (EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh faecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.
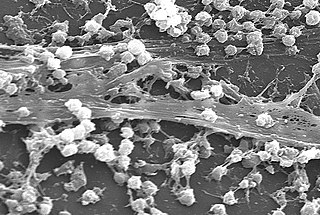
A biofilm comprises any syntrophic consortium of microorganisms in which cells stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric conglomeration of extracellular polysaccharides, proteins, lipids and DNA. Because they have three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes".

Campylobacter is a genus of Gram-negative bacteria. Campylobacter typically appear comma- or s-shaped, and are motile. Some Campylobacter species can infect humans, sometimes causing campylobacteriosis, a diarrhoeal disease in humans. Campylobacteriosis is usually self-limiting and antimicrobial treatment is often not required, except in severe cases or immunocompromised patients. The most known source for Campylobacter is poultry, but due to their diverse natural reservoir, Campylobacter spp. can also be transmitted via water. Other known sources of Campylobacter infections include food products, such as unpasteurised milk and contaminated fresh produce. Sometimes the source of infection can be direct contact with infected animals, which often carry Campylobacter asymptomatically. At least a dozen species of Campylobacter have been implicated in human disease, with C. jejuni (80–90%) and C. coli (5-10%) being the most common. C. jejuni is recognized as one of the main causes of bacterial foodborne disease in many developed countries. It is the number one cause of bacterial gastroentritis in Europe, with over 246,000 cases confirmed annually. C. jejuni infection can also cause bacteremia in immunocompromised people, while C. lari is a known cause of recurrent diarrhea in children. C. fetus can cause spontaneous abortions in cattle and sheep, and is an opportunistic pathogen in humans.

Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher order evolution while more significantly shifting perspectives on bacterial evolution.

Mycoplasma is a genus of bacteria that, like the other members of the class Mollicutes, lack a cell wall around their cell membranes. Peptidoglycan (murein) is absent. This characteristic makes them naturally resistant to antibiotics that target cell wall synthesis. They can be parasitic or saprotrophic. Several species are pathogenic in humans, including M. pneumoniae, which is an important cause of "walking" pneumonia and other respiratory disorders, and M. genitalium, which is believed to be involved in pelvic inflammatory diseases. Mycoplasma species are among the smallest organisms yet discovered, can survive without oxygen, and come in various shapes. For example, M. genitalium is flask-shaped, while M. pneumoniae is more elongated, many Mycoplasma species are coccoid. Hundreds of Mycoplasma species infect animals.
Virulence is a pathogen's or microorganism's ability to cause damage to a host.

Campylobacter jejuni is a species of pathogenic bacteria, one of the most common causes of food poisoning in Europe and in the US. The vast majority of cases occur as isolated events, not as part of recognized outbreaks. Active surveillance through the Foodborne Diseases Active Surveillance Network (FoodNet) indicates that about 20 cases are diagnosed each year for each 100,000 people in the US, while many more cases are undiagnosed or unreported; the CDC estimates a total of 1.5 million infections every year. The European Food Safety Authority reported 246,571 cases in 2018, and estimated approximately nine million cases of human campylobacteriosis per year in the European Union.

Dickeya dadantii is a gram-negative bacillus that belongs to the family Pectobacteriaceae. It was formerly known as Erwinia chrysanthemi but was reassigned as Dickeya dadantii in 2005. Members of this family are facultative anaerobes, able to ferment sugars to lactic acid, have nitrate reductase, but lack oxidases. Even though many clinical pathogens are part of the order Enterobacterales, most members of this family are plant pathogens. D. dadantii is a motile, nonsporing, straight rod-shaped cell with rounded ends. Cells range in size from 0.8 to 3.2 μm by 0.5 to 0.8 μm and are surrounded by numerous flagella (peritrichous).
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:

Food microbiology is the study of the microorganisms that inhabit, create, or contaminate food. This includes the study of microorganisms causing food spoilage; pathogens that may cause disease ; microbes used to produce fermented foods such as cheese, yogurt, bread, beer, and wine; and microbes with other useful roles, such as producing probiotics.

Bacteria are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.

Genetically modified bacteria were the first organisms to be modified in the laboratory, due to their simple genetics. These organisms are now used for several purposes, and are particularly important in producing large amounts of pure human proteins for use in medicine.

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.
In biology, a pathogen in the oldest and broadest sense, is any organism or agent that can produce disease. A pathogen may also be referred to as an infectious agent, or simply a germ.
The host–pathogen interaction is defined as how microbes or viruses sustain themselves within host organisms on a molecular, cellular, organismal or population level. This term is most commonly used to refer to disease-causing microorganisms although they may not cause illness in all hosts. Because of this, the definition has been expanded to how known pathogens survive within their host, whether they cause disease or not.

Genetic engineering is the science of manipulating genetic material of an organism. The first artificial genetic modification accomplished using biotechnology was transgenesis, the process of transferring genes from one organism to another, first accomplished by Herbert Boyer and Stanley Cohen in 1973. It was the result of a series of advancements in techniques that allowed the direct modification of the genome. Important advances included the discovery of restriction enzymes and DNA ligases, the ability to design plasmids and technologies like polymerase chain reaction and sequencing. Transformation of the DNA into a host organism was accomplished with the invention of biolistics, Agrobacterium-mediated recombination and microinjection. The first genetically modified animal was a mouse created in 1974 by Rudolf Jaenisch. In 1976 the technology was commercialised, with the advent of genetically modified bacteria that produced somatostatin, followed by insulin in 1978. In 1983 an antibiotic resistant gene was inserted into tobacco, leading to the first genetically engineered plant. Advances followed that allowed scientists to manipulate and add genes to a variety of different organisms and induce a range of different effects. Plants were first commercialized with virus resistant tobacco released in China in 1992. The first genetically modified food was the Flavr Savr tomato marketed in 1994. By 2010, 29 countries had planted commercialized biotech crops. In 2000 a paper published in Science introduced golden rice, the first food developed with increased nutrient value.
Campylobacter coli is a Gram-negative, microaerophilic, non-endospore-forming, S-shaped bacterial species within genus Campylobacter. In humans, it C. coli can cause campylobacteriosis, a diarrhoeal disease which is the most frequently reported foodborne illness in the European Union. C. coli grows slowly with an optimum temperature of 42 °C. When exposed to air for long periods, they become spherical or coccoid shaped.

Caenorhabditis elegans- microbe interactions are defined as any interaction that encompasses the association with microbes that temporarily or permanently live in or on the nematode C. elegans. The microbes can engage in a commensal, mutualistic or pathogenic interaction with the host. These include bacterial, viral, unicellular eukaryotic, and fungal interactions. In nature C. elegans harbours a diverse set of microbes. In contrast, C. elegans strains that are cultivated in laboratories for research purposes have lost the natural associated microbial communities and are commonly maintained on a single bacterial strain, Escherichia coli OP50. However, E. coli OP50 does not allow for reverse genetic screens because RNAi libraries have only been generated in strain HT115. This limits the ability to study bacterial effects on host phenotypes. The host microbe interactions of C. elegans are closely studied because of their orthologs in humans. Therefore, the better we understand the host interactions of C. elegans the better we can understand the host interactions within the human body.

Human interactions with microbes include both practical and symbolic uses of microbes, and negative interactions in the form of human, domestic animal, and crop diseases.

Milk borne diseases are any diseases caused by consumption of milk or dairy products infected or contaminated by pathogens. Milk borne diseases are one of the recurrent foodborne illnesses— between 1993 to 2012 over 120 outbreaks related to raw milk were recorded in the US with approximately 1,900 illnesses and 140 hospitalisations. With rich nutrients essential for growth and development such as proteins, lipids, carbohydrates, and vitamins in milk, pathogenic microorganisms are well nourished and are capable of rapid cell division and extensive population growth in this favourable environment. Common pathogens include bacteria, viruses, fungi, and parasites and among them, bacterial infection is the leading cause of milk borne diseases.
















