
Streptococcus is a genus of gram-positive coccus or spherical bacteria that belongs to the family Streptococcaceae, within the order Lactobacillales, in the phylum Bacillota. Cell division in streptococci occurs along a single axis, so as they grow, they tend to form pairs or chains that may appear bent or twisted. This differs from staphylococci, which divide along multiple axes, thereby generating irregular, grape-like clusters of cells. Most streptococci are oxidase-negative and catalase-negative, and many are facultative anaerobes.

Enterococcus is a large genus of lactic acid bacteria of the phylum Bacillota. Enterococci are gram-positive cocci that often occur in pairs (diplococci) or short chains, and are difficult to distinguish from streptococci on physical characteristics alone. Two species are common commensal organisms in the intestines of humans: E. faecalis (90–95%) and E. faecium (5–10%). Rare clusters of infections occur with other species, including E. casseliflavus, E. gallinarum, and E. raffinosus.

Lactococcus is a genus of lactic acid bacteria that were formerly included in the genus Streptococcus Group N1. They are known as homofermenters meaning that they produce a single product, lactic acid in this case, as the major or only product of glucose fermentation. Their homofermentative character can be altered by adjusting environmental conditions such as pH, glucose concentration, and nutrient limitation. They are gram-positive, catalase-negative, non-motile cocci that are found singly, in pairs, or in chains. The genus contains strains known to grow at or below 7˚C.

Escherichia is a genus of Gram-negative, non-spore-forming, facultatively anaerobic, rod-shaped bacteria from the family Enterobacteriaceae. In those species which are inhabitants of the gastrointestinal tracts of warm-blooded animals, Escherichia species provide a portion of the microbially derived vitamin K for their host. A number of the species of Escherichia are pathogenic. The genus is named after Theodor Escherich, the discoverer of Escherichia coli. Escherichia are facultative aerobes, with both aerobic and anaerobic growth, and an optimum temperature of 37 °C. Escherichia are usually motile by flagella, produce gas from fermentable carbohydrates, and do not decarboxylate lysine or hydrolyze arginine. Species include E. albertii, E. fergusonii, E. hermannii, E. marmotae and most notably, the model organism and clinically relevant E. coli. Shimwellia blattae and Pseudescherichia vulneris was formerly classified in this genus.

Enterobacter is a genus of common Gram-negative, facultatively anaerobic, rod-shaped, non-spore-forming bacteria of the family Enterobacteriaceae. It is the type genus of the order Enterobacterales. Several strains of these bacteria are pathogenic and cause opportunistic infections in immunocompromised hosts and in those who are on mechanical ventilation. The urinary and respiratory tracts are the most common sites of infection. The genus Enterobacter is a member of the coliform group of bacteria. It does not belong to the fecal coliforms group of bacteria, unlike Escherichia coli, because it is incapable of growth at 44.5 °C in the presence of bile salts. Some of them show quorum sensing properties.

Vancomycin-resistant Enterococcus, or vancomycin-resistant enterococci (VRE), are bacterial strains of the genus Enterococcus that are resistant to the antibiotic vancomycin.

Enterococcus faecalis – formerly classified as part of the group D Streptococcus system – is a Gram-positive, commensal bacterium inhabiting the gastrointestinal tracts of humans. Like other species in the genus Enterococcus, E. faecalis is found in healthy humans and can be used as a probiotic. The probiotic strains such as Symbioflor1 and EF-2001 are characterized by the lack of specific genes related to drug resistance and pathogenesis. As an opportunistic pathogen, E. faecalis can cause life-threatening infections, especially in the nosocomial (hospital) environment, where the naturally high levels of antibiotic resistance found in E. faecalis contribute to its pathogenicity. E. faecalis has been frequently found in reinfected, root canal-treated teeth in prevalence values ranging from 30% to 90% of the cases. Re-infected root canal-treated teeth are about nine times more likely to harbor E. faecalis than cases of primary infections.
Streptococcus bovis is a species of Gram-positive bacteria that in humans is associated with urinary tract infections, endocarditis, sepsis, and colorectal cancer. S. gallolyticus is commonly found in the alimentary tract of cattle, sheep, and other ruminants, and may cause ruminal acidosis or feedlot bloat. It is also associated with spontaneous bacterial peritonitis, a frequent complication occurring in patients affected by cirrhosis. Equivalence with Streptococcus equinus has been contested.
Gemella morbillorum is a species of bacteria within the genus Gemella. It is a facultative anaerobic Gram positive coccus usually preferring capnophilic or microaerophilic environments. From its discovery in 1917 until 1988, it was known as Streptococcus morbillorum. The name change followed closer examination with DNA filter hybridization which found it was very close to the species Gemella haemolysans.

Lactobacillales are an order of gram-positive, low-GC, acid-tolerant, generally nonsporulating, nonrespiring, either rod-shaped (bacilli) or spherical (cocci) bacteria that share common metabolic and physiological characteristics. These bacteria, usually found in decomposing plants and milk products, produce lactic acid as the major metabolic end product of carbohydrate fermentation, giving them the common name lactic acid bacteria (LAB).
Enterococcus faecium is a Gram-positive, gamma-hemolytic or non-hemolytic bacterium in the genus Enterococcus. It can be commensal in the gastrointestinal tract of humans and animals, but it may also be pathogenic, causing diseases such as neonatal meningitis or endocarditis.
Streptococcus equinus is a Gram-positive, nonhemolytic, nonpathogenic, lactic acid bacterium of the genus Streptococcus. It is the principal Streptococcus found in the alimentary canal of a horse, and makes up the majority of the bacterial flora in horse feces. Equivalence with Streptococcus bovis has been contested.
Enterococcus gallinarum is a species of Enterococcus. E. gallinarum demonstrates an inherent, low-level resistance to vancomycin. Resistance is due to a chromosomal gene, vanC, which encodes for a terminal D-alanine-D-serine instead of the usual D-alanine-D-alanine in cell wall peptidoglycan precursor proteins. That is a separate mechanism than the vancomycin resistance seen in VRE isolates of E. faecium and E. faecalis which is mediated by vanA or vanB. This species is known to cause clusters of infection, although it considered very rare. It is the only other known enterococcal species besides E. faecium and E. faecalis known to cause outbreaks and spread in hospitals.

Streptococcus iniae is a species of Gram-positive, sphere-shaped bacterium belonging to the genus Streptococcus. Since its isolation from an Amazon freshwater dolphin in the 1970s, S. iniae has emerged as a leading fish pathogen in aquaculture operations worldwide, resulting in over US$100M in annual losses. Since its discovery, S. iniae infections have been reported in at least 27 species of cultured or wild fish from around the world. Freshwater and saltwater fish including tilapia, red drum, hybrid striped bass, and rainbow trout are among those susceptible to infection by S. iniae. Infections in fish manifest as meningoencephalitis, skin lesions, and septicemia.
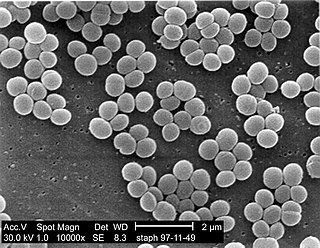
Staphylococcus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical (cocci), and form in grape-like clusters. Staphylococcus species are facultative anaerobic organisms.

Lancefield grouping is a system of classification that classifies catalase-negative Gram-positive cocci based on the carbohydrate composition of bacterial antigens found on their cell walls. The system, created by Rebecca Lancefield, was historically used to organize the various members of the family Streptococcaceae, which includes the genera Lactococcus and Streptococcus, but now is largely superfluous due to explosive growth in the number of streptococcal species identified since the 1970s. However, it has retained some clinical usefulness even after the taxonomic changes, and as of 2018, Lancefield designations are still often used to communicate medical microbiological test results.
Vagococcus fluvialis is a species of bacteria. The type strain of V. fluvialis is NCDO 2497. It rarely causes human infection. The only genetically proven case of V. fluvialis endocarditis was detected in the Cochin, India.
Enterococcus raffinosus is a bacterial species of the Gram-positive genus Enterococcus, named for its facultative anaerobic metabolism, including the ability to ferment the trisaccharide raffinose. This mesophilic microaerophile has optimal growth at 37ºC in Columbia Blood Medium. It has an ovoid morphology categorized as coccal with arrangement singly, in pairs, or short chains.
Enterococcus pseudoavium is a species of Enterococcus.
Methanogens are a group of microorganisms that produce methane as a byproduct of their metabolism. They play an important role in the digestive system of ruminants. The digestive tract of ruminants contains four major parts: rumen, reticulum, omasum and abomasum. The food with saliva first passes to the rumen for breaking into smaller particles and then moves to the reticulum, where the food is broken into further smaller particles. Any indigestible particles are sent back to the rumen for rechewing. The majority of anaerobic microbes assisting the cellulose breakdown occupy the rumen and initiate the fermentation process. The animal absorbs the fatty acids, vitamins and nutrient content on passing the partially digested food from the rumen to the omasum. This decreases the pH level and initiates the release of enzymes for further breakdown of the food which later passes to the abomasum to absorb remaining nutrients before excretion. This process takes about 9–12 hours.










