
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.
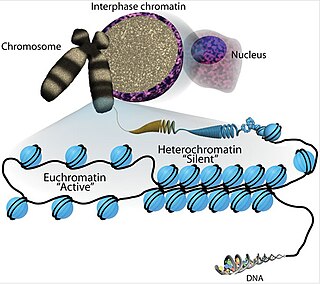
Euchromatin is a lightly packed form of chromatin that is enriched in genes, and is often under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic.
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.

Constitutive heterochromatin domains are regions of DNA found throughout the chromosomes of eukaryotes. The majority of constitutive heterochromatin is found at the pericentromeric regions of chromosomes, but is also found at the telomeres and throughout the chromosomes. In humans there is significantly more constitutive heterochromatin found on chromosomes 1, 9, 16, 19 and Y. Constitutive heterochromatin is composed mainly of high copy number tandem repeats known as satellite repeats, minisatellite and microsatellite repeats, and transposon repeats. In humans these regions account for about 200Mb or 6.5% of the total human genome, but their repeat composition makes them difficult to sequence, so only small regions have been sequenced.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.
Position-effect variegation (PEV) is a variegation caused by the silencing of a gene in some cells through its abnormal juxtaposition with heterochromatin via rearrangement or transposition. It is also associated with changes in chromatin conformation.

Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the CTCF gene. CTCF is involved in many cellular processes, including transcriptional regulation, insulator activity, V(D)J recombination and regulation of chromatin architecture.
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome. The field is analogous to genomics and proteomics, which are the study of the genome and proteome of a cell. Epigenetic modifications are reversible modifications on a cell's DNA or histones that affect gene expression without altering the DNA sequence. Epigenomic maintenance is a continuous process and plays an important role in stability of eukaryotic genomes by taking part in crucial biological mechanisms like DNA repair. Plant flavones are said to be inhibiting epigenomic marks that cause cancers. Two of the most characterized epigenetic modifications are DNA methylation and histone modification. Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis. The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.

Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or other DNA binding sites, and possibly define cistromes. ChIP also aims to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers. ChIP is crucial for the advancements in the field of epigenomics and learning more about epigenetic phenomena.
FAIRE-Seq is a method in molecular biology used for determining the sequences of DNA regions in the genome associated with regulatory activity. The technique was developed in the laboratory of Jason D. Lieb at the University of North Carolina, Chapel Hill. In contrast to DNase-Seq, the FAIRE-Seq protocol doesn't require the permeabilization of cells or isolation of nuclei, and can analyse any cell type. In a study of seven diverse human cell types, DNase-seq and FAIRE-seq produced strong cross-validation, with each cell type having 1-2% of the human genome as open chromatin.

Biomarkers of aging are biomarkers that could predict functional capacity at some later age better than chronological age. Stated another way, biomarkers of aging would give the true "biological age", which may be different from the chronological age.
Embryonic stem cells are capable of self-renewing and differentiating to the desired fate depending on their position in the body. Stem cell homeostasis is maintained through epigenetic mechanisms that are highly dynamic in regulating the chromatin structure as well as specific gene transcription programs. Epigenetics has been used to refer to changes in gene expression, which are heritable through modifications not affecting the DNA sequence.
Human herpes viruses, also known as HHVs, are part of a family of DNA viruses that cause several diseases in humans. One of the most notable functions of this virus family is their ability to enter a latent phase and lay dormant within animals for extended periods of time. The mechanism that controls this is very complex because expression of viral proteins during latency is decreased a great deal, meaning that the virus must have transcription of its genes repressed. There are many factors and mechanisms that control this process and epigenetics is one way this is accomplished. Epigenetics refers to persistent changes in expression patterns that are not caused by changes to the DNA sequence. This happens through mechanisms such as methylation and acetylation of histones, DNA methylation, and non-coding RNAs (ncRNA). Altering the acetylation of histones creates changes in expression by changing the binding affinity of histones to DNA, making it harder or easier for transcription machinery to access the DNA. Methyl and acetyl groups can also act as binding sites for transcription factors and enzymes that further modify histones or alter the DNA itself.
H3K27me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein.

Robin Campbell Allshire is Professor of Chromosome Biology at University of Edinburgh and a Wellcome Trust Principal Research Fellow. His research group at the Wellcome Trust Centre for Cell Biology focuses on the epigenetic mechanisms governing the assembly of specialised domains of chromatin and their transmission through cell division.

Thomas Jenuwein is a German scientist working in the fields of epigenetics, chromatin biology, gene regulation and genome function.
H3K9me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation at the 9th lysine residue of the histone H3 protein and is often associated with heterochromatin.
Human epigenome is the complete set of structural modifications of chromatin and chemical modifications of histones and nucleotides. These modifications affect according to cellular type and development status. Various studies show that epigenome depends on exogenous factors.
H3K36me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation at the 36th lysine residue of the histone H3 protein and often associated with gene bodies.
H4K20me is an epigenetic modification to the DNA packaging protein Histone H4. It is a mark that indicates the mono-methylation at the 20th lysine residue of the histone H4 protein. This mark can be di- and tri-methylated. It is critical for genome integrity including DNA damage repair, DNA replication and chromatin compaction.











