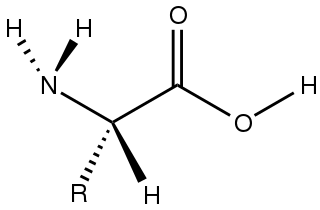
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life.

Alanine, or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side chain. Consequently it is classified as a nonpolar, aliphatic α-amino acid. Under biological conditions, it exists in its zwitterionic form with its amine group protonated and its carboxyl group deprotonated. It is non-essential to humans as it can be synthesized metabolically and does not need to be present in the diet. It is encoded by all codons starting with GC.

Proteinogenic amino acids are amino acids that are incorporated biosynthetically into proteins during translation. The word "proteinogenic" means "protein creating". Throughout known life, there are 22 genetically encoded (proteinogenic) amino acids, 20 in the standard genetic code and an additional 2 that can be incorporated by special translation mechanisms.

A codon table can be used to translate a genetic code into a sequence of amino acids. The standard genetic code is traditionally represented as an RNA codon table, because when proteins are made in a cell by ribosomes, it is messenger RNA (mRNA) that directs protein synthesis. The mRNA sequence is determined by the sequence of genomic DNA. In this context, the standard genetic code is referred to as translation table 1. It can also be represented in a DNA codon table. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5′-to-3′ direction. Different tables with alternate codons are used depending on the source of the genetic code, such as from a cell nucleus, mitochondrion, plastid, or hydrogenosome.

In biochemistry, non-coded or non-proteinogenic amino acids are distinct from the 22 proteinogenic amino acids, which are naturally encoded in the genome of organisms for the assembly of proteins. However, over 140 non-proteinogenic amino acids occur naturally in proteins and thousands more may occur in nature or be synthesized in the laboratory. Chemically synthesized amino acids can be called unnatural amino acids. Unnatural amino acids can be synthetically prepared from their native analogs via modifications such as amine alkylation, side chain substitution, structural bond extension cyclization, and isosteric replacements within the amino acid backbone. Many non-proteinogenic amino acids are important:
The yeast mitochondrial code is a genetic code used by the mitochondrial genome of yeasts, notably Saccharomyces cerevisiae, Candida glabrata, Hansenula saturnus, and Kluyveromyces thermotolerans.
The mold, protozoan, and coelenterate mitochondrial code and the mycoplasma/spiroplasma code is the genetic code used by various organisms, in some cases with slight variations, notably the use of UGA as a tryptophan codon rather than a stop codon.
GADV-protein world is a hypothetical stage of abiogenesis. GADV stands for the one letter codes of four amino acids, namely, glycine (G), alanine (A), aspartic acid (D) and valine (V), the main components of GADV proteins. In the GADV-protein world hypothesis, it is argued that the prebiotic chemistry before the emergence of genes involved a stage where GADV-proteins were able to pseudo-replicate. This hypothesis is contrary to the RNA world hypothesis.
The candidate division SR1 and gracilibacteria code is used in two groups of uncultivated bacteria found in marine and fresh-water environments and in the intestines and oral cavities of mammals among others. The difference to the standard and the bacterial code is that UGA represents an additional glycine codon and does not code for termination. A survey of many genomes with the codon assignment software Codetta, analyzed through the GTDB taxonomy system shows that this genetic code is limited to the Patescibacteria order BD1-5, not what are now termed Gracilibacteria, and that the SR1 genome assembly GCA_000350285.1 for which the table 25 code was originally defined is actually using the Absconditibacterales genetic code and has the associated three special recoding tRNAs. Thus this code may now be better named the "BD1-5 code".
The Blepharisma nuclear code is a genetic code found in the nuclei of Blepharisma.
The chlorophycean mitochondrial code is a genetic code found in the mitochondria of Chlorophyceae.
The trematode mitochondrial code is a genetic code found in the mitochondria of Trematoda.
The pachysolen tannophilus nuclear code is a genetic code found in the ascomycete fungus Pachysolen tannophilus.
The karyorelictid nuclear code is a genetic code used by the nuclear genome of the Karyorelictea ciliate Parduczia sp. This code, along with translation tables 28 and 31, is remarkable in that every one of the 64 possible codons can be a sense codon. Translation termination probably relies on context, specifically proximity to the poly(A) tail.
The Condylostoma nuclear code is a genetic code used by the nuclear genome of the heterotrich ciliate Condylostoma magnum. This code, along with translation tables 27 and 31, is remarkable in that every one of the 64 possible codons can be a sense codon. Experimental evidence suggests that translation termination relies on context, specifically proximity to the poly(A) tail. Near such a tail, PABP could help terminate the protein by recruiting eRF1 and eRF3 to prevent the cognate tRNA from binding.
The Mesodinium nuclear code is a genetic code used by the nuclear genome of the ciliates Mesodinium and Myrionecta.
The Blastocrithidia nuclear code is a genetic code used by the nuclear genome of the trypanosomatid genus Blastocrithidia. This code, along with translation tables 27 and 28, is remarkable in that every one of the 64 possible codons can be a sense codon.
Low complexity regions (LCRs) in protein sequences, also defined in some contexts as compositionally biased regions (CBRs), are regions in protein sequences that differ from the composition and complexity of most proteins that is normally associated with globular structure. LCRs have different properties from normal regions regarding structure, function and evolution.
The QTY Code is a design method to transform membrane proteins that are intrinsically insoluble in water into variants with water solubility, while retaining their structure and function.
Phoratoxins are a group of peptide toxins that belong to the family of thionins, a subdivision of small plant toxins. Phoratoxins are proteins present in the leaves and branches of the Phoradendron, commonly known as the American variant of the mistletoe, a plant commonly used as decoration during the festive season. The berries of the mistletoe do not contain phoratoxins, making them less toxic compared to other parts of the plant. The toxicity of the mistletoe is dependent on the host tree, since mistletoe is known to be a semi-parasite. The host tree provides fixed inorganic nitrogen compounds necessary for the mistletoe to synthesize phoratoxins.



