Related Research Articles

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.

Electron-capture dissociation (ECD) is a method of fragmenting gas-phase ions for structure elucidation of peptides and proteins in tandem mass spectrometry. It is one of the most widely used techniques for activation and dissociation of mass selected precursor ion in MS/MS. It involves the direct introduction of low-energy electrons to trapped gas-phase ions.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.
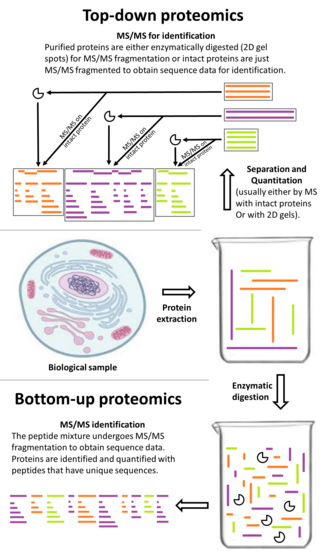
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.
Robert Graham Cooks is the Henry Bohn Hass Distinguished Professor of Chemistry in the Aston Laboratories for Mass Spectrometry at Purdue University. He is an ISI Highly Cited Chemist, with over 1,000 publications and an H-index of 144.
Michael L. Gross is Professor of Chemistry, Medicine, and Immunology, at Washington University in St. Louis. He was formerly Professor of Chemistry at the University of Nebraska-Lincoln from 1968–1994. He is recognized for his contributions to the field of mass spectrometry and ion chemistry. He is credited with the discovery of distonic ions, chemical species containing a radical and an ionic site on different atoms of the same molecule.
Joshua Coon is a professor of chemistry and biomolecular chemistry and the inaugural holder of the Thomas and Margaret Pyle Chair at the University of Wisconsin–Madison, and an affiliate of the Morgridge Institute for Research.

Neil L. Kelleher is the Walter and Mary Elizabeth Glass Professor of Chemistry, Molecular Biosciences, and Medicine at Northwestern University. His research focuses on mass spectrometry, primarily its application to proteomics. He is known mainly for top-down proteomics and the development of the fragmentation technique of electron-capture dissociation with Roman Zubarev while in Fred McLafferty's lab at Cornell University.

Renato Zenobi is a Swiss chemist. He is Professor of Chemistry at ETH Zurich. Throughout his career, Zenobi has contributed to the field of analytical chemistry.

Albert J.R. Heck is a Dutch scientist and professor at Utrecht University, the Netherlands in the field of mass spectrometry and proteomics. He is known for his work on technologies to study proteins in their natural environment, with the aim to understand their biological function. Albert Heck was awarded the Spinoza Prize in 2017, the highest scientific award in the Netherlands.

Catherine Clarke Fenselau is an American scientist who was the first trained mass spectrometrist on the faculty of an American medical school; she joined Johns Hopkins School of Medicine in 1968. She specializes in biomedical applications of mass spectrometry. She has been recognized as an outstanding scientist in the field of bioanalytical chemistry because of her work using mass spectrometry to study biomolecules.
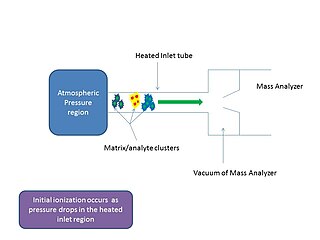
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.
Kristina Håkansson is an analytical chemist known for her contribution in Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry for biomolecular identification and structural characterization. Currently, she holds the position of Professor of Chemistry at University of Michigan. Her research focuses on mass spectrometry, primarily identification and characterization of protein posttranslational modifications by complementary fragmentation techniques such as electron-capture dissociation (ECD)/negative ion ECD (niECD) and infrared multiphoton dissociation (IRMPD) at low (femtomole) levels.
Peter Nemes is a Hungarian-American chemist, who is active in the fields of bioanalytical chemistry, mass spectrometry, cell/developmental biology, neuroscience, and biochemistry.
Ying Ge is a Chinese-American chemist who is a Professor of Cell and Regenerative Biology at the University of Wisconsin–Madison. Her research considers the molecular mechanisms that underpin cardiac disease. She has previously served on the board of directors of the American Society for Mass Spectrometry. In 2020 Ge was named on the Analytical Scientist Power List.

Ron M.A. Heeren is a Dutch scientist in mass spectrometry imaging. He is currently a distinguished professor at Maastricht University and the scientific director of the Multimodal Molecular Imaging Institute (M4I), where he heads the division of Imaging Mass Spectrometry.
Catherine E. Costello is the William Fairfield Warren distinguished professor in the department of biochemistry, Cell Biology and Genomics, and the director of the Center for Biomedical Mass Spectrometry at the Boston University School of Medicine.
Lingjun Li is a Professor in the School of Pharmacy and Department of Chemistry at University of Wisconsin-Madison. She develops mass spectrometry based tools to study neuropeptides, peptide hormones and neurotransmitters.
Hilkka Inkeri Kenttämaa is a researcher in organic and bioorganic mass spectrometry, and the Frank Brown Endowed Distinguished Professor of Chemistry at Purdue University. She is a pioneer in distonic radical cation research and laser-induced acoustic desorption.
Yu-Ju Chen is a Taiwanese proteomics research scientist, who leads international projects in proteogenomics.
References
- ↑ "Pushing the Boundaries of Mass Spectrometry Design and Use". Accelerating Proteomics. 2013-06-12. Retrieved 2022-06-08.
- 1 2 "HOME 6TH FORM ALUMNI PROFESSOR HELEN J. COOPER".
- 1 2 "Professor Helen Cooper - 2022 Analytical Division open Award: Theophilus Redwood Award winner". Royal Society of Chemistry. Retrieved 2022-06-08.
- 1 2 "Professor Helen Cooper". Rosalind Franklin Institute. Retrieved 2022-06-08.
- ↑ "Extract from IMMS: The Next Five Years - Prof. Helen J. Cooper". www.owlstonemedical.com. Retrieved 2022-06-08.
- 1 2 "Professor Helen Cooper". University of Birmingham. Retrieved 2022-06-08.
- ↑ "Editors & Editorial Board". Journal of the American Society for Mass Spectrometry. Retrieved 2024-04-28.
- ↑ "Our 2022 Research & Innovation Prize winners". Royal Society of Chemistry. Retrieved 2022-06-08.