
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.
Fourier-transform ion cyclotron resonance mass spectrometry is a type of mass analyzer (or mass spectrometer) for determining the mass-to-charge ratio (m/z) of ions based on the cyclotron frequency of the ions in a fixed magnetic field. The ions are trapped in a Penning trap (a magnetic field with electric trapping plates), where they are excited (at their resonant cyclotron frequencies) to a larger cyclotron radius by an oscillating electric field orthogonal to the magnetic field. After the excitation field is removed, the ions are rotating at their cyclotron frequency in phase (as a "packet" of ions). These ions induce a charge (detected as an image current) on a pair of electrodes as the packets of ions pass close to them. The resulting signal is called a free induction decay (FID), transient or interferogram that consists of a superposition of sine waves. The useful signal is extracted from this data by performing a Fourier transform to give a mass spectrum.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy-absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.
Infrared multiple photon dissociation (IRMPD) is a technique used in mass spectrometry to fragment molecules in the gas phase usually for structural analysis of the original (parent) molecule.

The history of mass spectrometry has its roots in physical and chemical studies regarding the nature of matter. The study of gas discharges in the mid 19th century led to the discovery of anode and cathode rays, which turned out to be positive ions and electrons. Improved capabilities in the separation of these positive ions enabled the discovery of stable isotopes of the elements. The first such discovery was with the element neon, which was shown by mass spectrometry to have at least two stable isotopes: 20Ne and 22Ne. Mass spectrometers were used in the Manhattan Project for the separation of isotopes of uranium necessary to create the atomic bomb.
Hydrogen–deuterium exchange is a chemical reaction in which a covalently bonded hydrogen atom is replaced by a deuterium atom, or vice versa. It can be applied most easily to exchangeable protons and deuterons, where such a transformation occurs in the presence of a suitable deuterium source, without any catalyst. The use of acid, base or metal catalysts, coupled with conditions of increased temperature and pressure, can facilitate the exchange of non-exchangeable hydrogen atoms, so long as the substrate is robust to the conditions and reagents employed. This often results in perdeuteration: hydrogen-deuterium exchange of all non-exchangeable hydrogen atoms in a molecule.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.
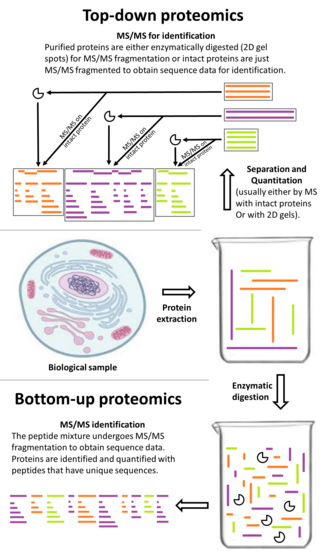
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.
Michael L. Gross is Professor of Chemistry, Medicine, and Immunology, at Washington University in St. Louis. He was formerly Professor of Chemistry at the University of Nebraska-Lincoln from 1968–1994. He is recognized for his contributions to the field of mass spectrometry and ion chemistry. He is credited with the discovery of distonic ions, chemical species containing a radical and an ionic site on different atoms of the same molecule.

Ambient ionization is a form of ionization in which ions are formed in an ion source outside the mass spectrometer without sample preparation or separation. Ions can be formed by extraction into charged electrospray droplets, thermally desorbed and ionized by chemical ionization, or laser desorbed or ablated and post-ionized before they enter the mass spectrometer.

In mass spectrometry, fragmentation is the dissociation of energetically unstable molecular ions formed from passing the molecules mass spectrum. These reactions are well documented over the decades and fragmentation patterns are useful to determine the molar weight and structural information of unknown molecules. Fragmentation that occurs in tandem mass spectrometry experiments has been a recent focus of research, because this data helps facilitate the identification of molecules.
Roman A. Zubarev is a professor of medicinal proteomics in the Department of Medical Biochemistry and Biophysics at the Karolinska Institutet. His research focuses on the use of mass spectrometry in biology and medicine.

Collision-induced dissociation (CID), also known as collisionally activated dissociation (CAD), is a mass spectrometry technique to induce fragmentation of selected ions in the gas phase. The selected ions are usually accelerated by applying an electrical potential to increase the ion kinetic energy and then allowed to collide with neutral molecules. In the collision, some of the kinetic energy is converted into internal energy which results in bond breakage and the fragmentation of the molecular ion into smaller fragments. These fragment ions can then be analyzed by tandem mass spectrometry.

Atmospheric pressure photoionization (APPI) is a soft ionization method used in mass spectrometry (MS) usually coupled to liquid chromatography (LC). Molecules are ionized using a vacuum ultraviolet (VUV) light source operating at atmospheric pressure, either by direct absorption followed by electron ejection or through ionization of a dopant molecule that leads to chemical ionization of target molecules. The sample is usually a solvent spray that is vaporized by nebulization and heat. The benefit of APPI is that it ionizes molecules across a broad range of polarity and is particularly useful for ionization of low polarity molecules for which other popular ionization methods such as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) are less suitable. It is also less prone to ion suppression and matrix effects compared to ESI and APCI and typically has a wide linear dynamic range. The application of APPI with LC/MS is commonly used for analysis of petroleum compounds, pesticides, steroids, and drug metabolites lacking polar functional groups and is being extensively deployed for ambient ionization particularly for explosives detection in security applications.
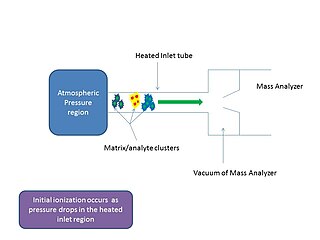
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.
Kristina Håkansson is an analytical chemist known for her contribution in Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry for biomolecular identification and structural characterization. Currently, she holds the position of Professor of Chemistry at University of Michigan. Her research focuses on mass spectrometry, primarily identification and characterization of protein posttranslational modifications by complementary fragmentation techniques such as electron-capture dissociation (ECD)/negative ion ECD (niECD) and infrared multiphoton dissociation (IRMPD) at low (femtomole) levels.
Ying Ge is a Chinese-American chemist who is a Professor of Cell and Regenerative Biology at the University of Wisconsin–Madison. Her research considers the molecular mechanisms that underpin cardiac disease. She has previously served on the board of directors of the American Society for Mass Spectrometry. In 2020 Ge was named on the Analytical Scientist Power List.
















