Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

Electron ionization is an ionization method in which energetic electrons interact with solid or gas phase atoms or molecules to produce ions. EI was one of the first ionization techniques developed for mass spectrometry. However, this method is still a popular ionization technique. This technique is considered a hard ionization method, since it uses highly energetic electrons to produce ions. This leads to extensive fragmentation, which can be helpful for structure determination of unknown compounds. EI is the most useful for organic compounds which have a molecular weight below 600 amu. Also, several other thermally stable and volatile compounds in solid, liquid and gas states can be detected with the use of this technique when coupled with various separation methods.

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.

Gas chromatography–mass spectrometry (GC–MS) is an analytical method that combines the features of gas-chromatography and mass spectrometry to identify different substances within a test sample. Applications of GC–MS include drug detection, fire investigation, environmental analysis, explosives investigation, food and flavor analysis, and identification of unknown samples, including that of material samples obtained from planet Mars during probe missions as early as the 1970s. GC–MS can also be used in airport security to detect substances in luggage or on human beings. Additionally, it can identify trace elements in materials that were previously thought to have disintegrated beyond identification. Like liquid chromatography–mass spectrometry, it allows analysis and detection even of tiny amounts of a substance.

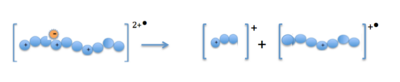
Electron-capture dissociation (ECD) is a method of fragmenting gas-phase ions for structure elucidation of peptides and proteins in tandem mass spectrometry. It is one of the most widely used techniques for activation and dissociation of mass selected precursor ion in MS/MS. It involves the direct introduction of low-energy electrons to trapped gas-phase ions.
Hydrogen–deuterium exchange is a chemical reaction in which a covalently bonded hydrogen atom is replaced by a deuterium atom, or vice versa. It can be applied most easily to exchangeable protons and deuterons, where such a transformation occurs in the presence of a suitable deuterium source, without any catalyst. The use of acid, base or metal catalysts, coupled with conditions of increased temperature and pressure, can facilitate the exchange of non-exchangeable hydrogen atoms, so long as the substrate is robust to the conditions and reagents employed. This often results in perdeuteration: hydrogen-deuterium exchange of all non-exchangeable hydrogen atoms in a molecule.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.

Time-of-flight mass spectrometry (TOFMS) is a method of mass spectrometry in which an ion's mass-to-charge ratio is determined by a time of flight measurement. Ions are accelerated by an electric field of known strength. This acceleration results in an ion having the same kinetic energy as any other ion that has the same charge. The velocity of the ion depends on the mass-to-charge ratio. The time that it subsequently takes for the ion to reach a detector at a known distance is measured. This time will depend on the velocity of the ion, and therefore is a measure of its mass-to-charge ratio. From this ratio and known experimental parameters, one can identify the ion.
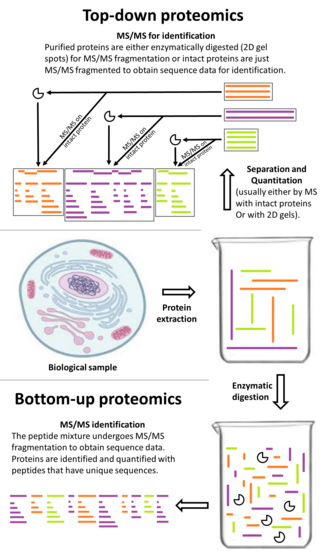
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.

Laser spray ionization refers to one of several methods for creating ions using a laser interacting with a spray of neutral particles or ablating material to create a plume of charged particles. The ions thus formed can be separated by m/z with mass spectrometry. Laser spray is one of several ion sources that can be coupled with liquid chromatography-mass spectrometry for the detection of larger molecules.

A triple quadrupole mass spectrometer (TQMS), is a tandem mass spectrometer consisting of two quadrupole mass analyzers in series, with a (non-mass-resolving) radio frequency (RF)–only quadrupole between them to act as a cell for collision-induced dissociation. This configuration is often abbreviated QqQ, here Q1q2Q3.
A hybrid mass spectrometer is a device for tandem mass spectrometry that consists of a combination of two or more m/z separation devices of different types.
Electron capture ionization is the ionization of a gas phase atom or molecule by attachment of an electron to create an ion of the form . The reaction is

Selected reaction monitoring (SRM), also called multiple reaction monitoring (MRM), is a method used in tandem mass spectrometry in which an ion of a particular mass is selected in the first stage of a tandem mass spectrometer and an ion product of a fragmentation reaction of the precursor ions is selected in the second mass spectrometer stage for detection.

The linear ion trap (LIT) is a type of ion trap mass spectrometer.

In mass spectrometry, fragmentation is the dissociation of energetically unstable molecular ions formed from passing the molecules mass spectrum. These reactions are well documented over the decades and fragmentation patterns are useful to determine the molar weight and structural information of unknown molecules. Fragmentation that occurs in tandem mass spectrometry experiments has been a recent focus of research, because this data helps facilitate the identification of molecules.
Joshua Coon is a professor of chemistry and biomolecular chemistry and the inaugural holder of the Thomas and Margaret Pyle Chair at the University of Wisconsin–Madison, and an affiliate of the Morgridge Institute for Research.

Collision-induced dissociation (CID), also known as collisionally activated dissociation (CAD), is a mass spectrometry technique to induce fragmentation of selected ions in the gas phase. The selected ions are usually accelerated by applying an electrical potential to increase the ion kinetic energy and then allowed to collide with neutral molecules. In the collision, some of the kinetic energy is converted into internal energy which results in bond breakage and the fragmentation of the molecular ion into smaller fragments. These fragment ions can then be analyzed by tandem mass spectrometry.
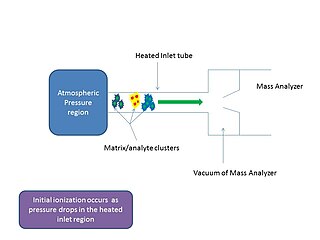
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.