Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

An ion source is a device that creates atomic and molecular ions. Ion sources are used to form ions for mass spectrometers, optical emission spectrometers, particle accelerators, ion implanters and ion engines.

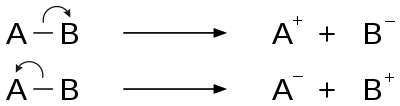
Electron ionization is an ionization method in which energetic electrons interact with solid or gas phase atoms or molecules to produce ions. EI was one of the first ionization techniques developed for mass spectrometry. However, this method is still a popular ionization technique. This technique is considered a hard ionization method, since it uses highly energetic electrons to produce ions. This leads to extensive fragmentation, which can be helpful for structure determination of unknown compounds. EI is the most useful for organic compounds which have a molecular weight below 600 amu. Also, several other thermally stable and volatile compounds in solid, liquid and gas states can be detected with the use of this technique when coupled with various separation methods.

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.

Chemical ionization (CI) is a soft ionization technique used in mass spectrometry. This was first introduced by Burnaby Munson and Frank H. Field in 1966. This technique is a branch of gaseous ion-molecule chemistry. Reagent gas molecules are ionized by electron ionization to form reagent ions, which subsequently react with analyte molecules in the gas phase to create analyte ions for analysis by mass spectrometry. Negative chemical ionization (NCI), charge-exchange chemical ionization, atmospheric-pressure chemical ionization (APCI) and atmospheric pressure photoionization (APPI) are some of the common variants of the technique. CI mass spectrometry finds general application in the identification, structure elucidation and quantitation of organic compounds as well as some utility in biochemical analysis. Samples to be analyzed must be in vapour form, or else, must be vapourized before introduction into the source.
Infrared multiple photon dissociation (IRMPD) is a technique used in mass spectrometry to fragment molecules in the gas phase usually for structural analysis of the original (parent) molecule.

Electron-capture dissociation (ECD) is a method of fragmenting gas-phase ions for structure elucidation of peptides and proteins in tandem mass spectrometry. It is one of the most widely used techniques for activation and dissociation of mass selected precursor ion in MS/MS. It involves the direct introduction of low-energy electrons to trapped gas-phase ions.
Gas phase ion chemistry is a field of science encompassed within both chemistry and physics. It is the science that studies ions and molecules in the gas phase, most often enabled by some form of mass spectrometry. By far the most important applications for this science is in studying the thermodynamics and kinetics of reactions. For example, one application is in studying the thermodynamics of the solvation of ions. Ions with small solvation spheres of 1, 2, 3... solvent molecules can be studied in the gas phase and then extrapolated to bulk solution.
Hydrogen–deuterium exchange is a chemical reaction in which a covalently bonded hydrogen atom is replaced by a deuterium atom, or vice versa. It can be applied most easily to exchangeable protons and deuterons, where such a transformation occurs in the presence of a suitable deuterium source, without any catalyst. The use of acid, base or metal catalysts, coupled with conditions of increased temperature and pressure, can facilitate the exchange of non-exchangeable hydrogen atoms, so long as the substrate is robust to the conditions and reagents employed. This often results in perdeuteration: hydrogen-deuterium exchange of all non-exchangeable hydrogen atoms in a molecule.
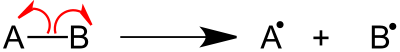
Alpha-cleavage (α-cleavage) in organic chemistry refers to the act of breaking the carbon-carbon bond adjacent to the carbon bearing a specified functional group.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.
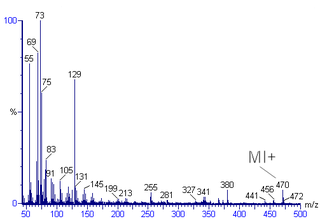
Mass spectral interpretation is the method employed to identify the chemical formula, characteristic fragment patterns and possible fragment ions from the mass spectra. Mass spectra is a plot of relative abundance against mass-to-charge ratio. It is commonly used for the identification of organic compounds from electron ionization mass spectrometry. Organic chemists obtain mass spectra of chemical compounds as part of structure elucidation and the analysis is part of many organic chemistry curricula.
Chemi-ionization is the formation of an ion through the reaction of a gas phase atom or molecule with another atom or molecule when the collision energy is below the energy required to ionize the reagents. The reaction may involve a reagent in an excited state or may result in the formation of a new chemical bond. Chemi-ionization can proceed through the Penning, associative, dissociative or rearrangement ionization reactions. Includes reactions that produce a free electron or a pair of ions.

A triple quadrupole mass spectrometer (TQMS), is a tandem mass spectrometer consisting of two quadrupole mass analyzers in series, with a (non-mass-resolving) radio frequency (RF)–only quadrupole between them to act as a cell for collision-induced dissociation. This configuration is often abbreviated QqQ, here Q1q2Q3.
Electron capture ionization is the ionization of a gas phase atom or molecule by attachment of an electron to create an ion of the form . The reaction is
Unimolecular ion decomposition is the fragmentation of a gas phase ion in a reaction with a molecularity of one. Ions with sufficient internal energy may fragment in a mass spectrometer, which in some cases may degrade the mass spectrometer performance, but in other cases, such as tandem mass spectrometry, the fragmentation can reveal information about the structure of the ion.

Collision-induced dissociation (CID), also known as collisionally activated dissociation (CAD), is a mass spectrometry technique to induce fragmentation of selected ions in the gas phase. The selected ions are usually accelerated by applying an electrical potential to increase the ion kinetic energy and then allowed to collide with neutral molecules. In the collision, some of the kinetic energy is converted into internal energy which results in bond breakage and the fragmentation of the molecular ion into smaller fragments. These fragment ions can then be analyzed by tandem mass spectrometry.

Atmospheric pressure photoionization (APPI) is a soft ionization method used in mass spectrometry (MS) usually coupled to liquid chromatography (LC). Molecules are ionized using a vacuum ultraviolet (VUV) light source operating at atmospheric pressure, either by direct absorption followed by electron ejection or through ionization of a dopant molecule that leads to chemical ionization of target molecules. The sample is usually a solvent spray that is vaporized by nebulization and heat. The benefit of APPI is that it ionizes molecules across a broad range of polarity and is particularly useful for ionization of low polarity molecules for which other popular ionization methods such as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) are less suitable. It is also less prone to ion suppression and matrix effects compared to ESI and APCI and typically has a wide linear dynamic range. The application of APPI with LC/MS is commonly used for analysis of petroleum compounds, pesticides, steroids, and drug metabolites lacking polar functional groups and is being extensively deployed for ambient ionization particularly for explosives detection in security applications.

Gary Siuzdak is an American chemist best known for his work in the field of metabolomics, activity metabolomics, and mass spectrometry. His lab discovered indole-3-propionic acid as a gut bacteria derived metabolite in 2009. He is currently the Professor and Director of The Center for Metabolomics and Mass Spectrometry at Scripps Research in La Jolla, California. Siuzdak has also made contributions to virus analysis, viral structural dynamics, as well as developing mass spectrometry imaging technology using nanostructured surfaces. The Siuzdak lab is also responsible for creating the research tools eXtensible Computational Mass Spectrometry (XCMS), METLIN, METLIN Neutral Loss and Q-MRM. As of January 2021, the XCMS/METLIN platform has over 50,000 registered users.