
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

A mass spectrum is a histogram plot of intensity vs. mass-to-charge ratio (m/z) in a chemical sample, usually acquired using an instrument called a mass spectrometer. Not all mass spectra of a given substance are the same; for example, some mass spectrometers break the analyte molecules into fragments; others observe the intact molecular masses with little fragmentation. A mass spectrum can represent many different types of information based on the type of mass spectrometer and the specific experiment applied. Common fragmentation processes for organic molecules are the McLafferty rearrangement and alpha cleavage. Straight chain alkanes and alkyl groups produce a typical series of peaks: 29 (CH3CH2+), 43 (CH3CH2CH2+), 57 (CH3CH2CH2CH2+), 71 (CH3CH2CH2CH2CH2+) etc.

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.

Gas chromatography–mass spectrometry (GC–MS) is an analytical method that combines the features of gas-chromatography and mass spectrometry to identify different substances within a test sample. Applications of GC–MS include drug detection, fire investigation, environmental analysis, explosives investigation, food and flavor analysis, and identification of unknown samples, including that of material samples obtained from planet Mars during probe missions as early as the 1970s. GC–MS can also be used in airport security to detect substances in luggage or on human beings. Additionally, it can identify trace elements in materials that were previously thought to have disintegrated beyond identification. Like liquid chromatography–mass spectrometry, it allows analysis and detection even of tiny amounts of a substance.

An ion trap is a combination of electric and/or magnetic fields used to capture charged particles — known as ions — often in a system isolated from an external environment. Atomic and molecular ion traps have a number of applications in physics and chemistry such as precision mass spectrometry, improved atomic frequency standards, and quantum computing. In comparison to neutral atom traps, ion traps have deeper trapping potentials that do not depend on the internal electronic structure of a trapped ion. This makes ion traps more suitable for the study of light interactions with single atomic systems. The two most popular types of ion traps are the Penning trap, which forms a potential via a combination of static electric and magnetic fields, and the Paul trap which forms a potential via a combination of static and oscillating electric fields.

In mass spectrometry, the quadrupole mass analyzer is a type of mass analyzer originally conceived by Nobel laureate Wolfgang Paul and his student Helmut Steinwedel. As the name implies, it consists of four cylindrical rods, set parallel to each other. In a quadrupole mass spectrometer (QMS) the quadrupole is the mass analyzer – the component of the instrument responsible for selecting sample ions based on their mass-to-charge ratio (m/z). Ions are separated in a quadrupole based on the stability of their trajectories in the oscillating electric fields that are applied to the rods.
Fourier-transform ion cyclotron resonance mass spectrometry is a type of mass analyzer (or mass spectrometer) for determining the mass-to-charge ratio (m/z) of ions based on the cyclotron frequency of the ions in a fixed magnetic field. The ions are trapped in a Penning trap (a magnetic field with electric trapping plates), where they are excited (at their resonant cyclotron frequencies) to a larger cyclotron radius by an oscillating electric field orthogonal to the magnetic field. After the excitation field is removed, the ions are rotating at their cyclotron frequency in phase (as a "packet" of ions). These ions induce a charge (detected as an image current) on a pair of electrodes as the packets of ions pass close to them. The resulting signal is called a free induction decay (FID), transient or interferogram that consists of a superposition of sine waves. The useful signal is extracted from this data by performing a Fourier transform to give a mass spectrum.

Liquid chromatography–mass spectrometry (LC–MS) is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography with the mass analysis capabilities of mass spectrometry (MS). Coupled chromatography – MS systems are popular in chemical analysis because the individual capabilities of each technique are enhanced synergistically. While liquid chromatography separates mixtures with multiple components, mass spectrometry provides spectral information that may help to identify each separated component. MS is not only sensitive, but provides selective detection, relieving the need for complete chromatographic separation. LC–MS is also appropriate for metabolomics because of its good coverage of a wide range of chemicals. This tandem technique can be used to analyze biochemical, organic, and inorganic compounds commonly found in complex samples of environmental and biological origin. Therefore, LC–MS may be applied in a wide range of sectors including biotechnology, environment monitoring, food processing, and pharmaceutical, agrochemical, and cosmetic industries. Since the early 2000s, LC–MS has also begun to be used in clinical applications.

Ion mobility spectrometry (IMS) It is a method of conducting analytical research that separates and identifies ionized molecules present in the gas phase based on the mobility of the molecules in a carrier buffer gas. Even though it is used extensively for military or security objectives, such as detecting drugs and explosives, the technology also has many applications in laboratory analysis, including studying small and big biomolecules. IMS instruments are extremely sensitive stand-alone devices, but are often coupled with mass spectrometry, gas chromatography or high-performance liquid chromatography in order to achieve a multi-dimensional separation. They come in various sizes, ranging from a few millimetres to several metres depending on the specific application, and are capable of operating under a broad range of conditions. IMS instruments such as microscale high-field asymmetric-waveform ion mobility spectrometry can be palm-portable for use in a range of applications including volatile organic compound (VOC) monitoring, biological sample analysis, medical diagnosis and food quality monitoring. Systems operated at higher pressure are often accompanied by elevated temperature, while lower pressure systems (1–20 hPa) do not require heating.

Atmospheric pressure chemical ionization (APCI) is an ionization method used in mass spectrometry which utilizes gas-phase ion-molecule reactions at atmospheric pressure (105 Pa), commonly coupled with high-performance liquid chromatography (HPLC). APCI is a soft ionization method similar to chemical ionization where primary ions are produced on a solvent spray. The main usage of APCI is for polar and relatively less polar thermally stable compounds with molecular weight less than 1500 Da. The application of APCI with HPLC has gained a large popularity in trace analysis detection such as steroids, pesticides and also in pharmacology for drug metabolites.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.

Ion mobility spectrometry–mass spectrometry (IMS-MS) is an analytical chemistry method that separates gas phase ions based on their interaction with a collision gas and their masses. In the first step, the ions are separated according to their mobility through a buffer gas on a millisecond timescale using an ion mobility spectrometer. The separated ions are then introduced into a mass analyzer in a second step where their mass-to-charge ratios can be determined on a microsecond timescale. The effective separation of analytes achieved with this method makes it widely applicable in the analysis of complex samples such as in proteomics and metabolomics.
Label-free quantification is a method in mass spectrometry that aims to determine the relative amount of proteins in two or more biological samples. Unlike other methods for protein quantification, label-free quantification does not use a stable isotope containing compound to chemically bind to and thus label the protein.
High-field asymmetric-waveform ion mobility spectrometry is an ion mobility spectrometry technique in which ions at atmospheric pressure are separated by the application of a high-voltage asymmetric waveform at radio frequency (RF) combined with a static (DC) waveform applied between two electrodes. Depending on the ratio of the high-field and low-field mobility of the ion, it will migrate toward one or the other electrode. Only ions with specific mobility will pass through the device.

A triple quadrupole mass spectrometer (TQMS), is a tandem mass spectrometer consisting of two quadrupole mass analyzers in series, with a (non-mass-resolving) radio frequency (RF)–only quadrupole between them to act as a cell for collision-induced dissociation. This configuration is often abbreviated QqQ, here Q1q2Q3.
A hybrid mass spectrometer is a device for tandem mass spectrometry that consists of a combination of two or more m/z separation devices of different types.

The linear ion trap (LIT) is a type of ion trap mass spectrometer.

A miniature mass spectrometer (MMS) is a type of mass spectrometer (MS) which has small size and weight and can be understood as a portable or handheld device. Current lab-scale mass spectrometers however, usually weigh hundreds of pounds and can cost on the range from thousands to millions of dollars. One purpose of producing MMS is for in situ analysis. This in situ analysis can lead to much simpler mass spectrometer operation such that non-technical personnel like physicians at the bedside, firefighters in a burning factory, food safety inspectors in a warehouse, or airport security at airport checkpoints, etc. can analyze samples themselves saving the time, effort, and cost of having the sample run by a trained MS technician offsite. Although, reducing the size of MS can lead to a poorer performance of the instrument versus current analytical laboratory standards, MMS is designed to maintain sufficient resolutions, detection limits, accuracy, and especially the capability of automatic operation. These features are necessary for the specific in-situ applications of MMS mentioned above.
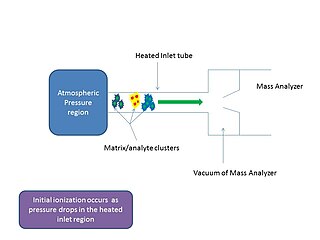
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.
SCIEX is a manufacturer of mass spectrometry instrumentation used in biomedical and environmental applications. Originally started by scientists from the University of Toronto Institute for Aerospace Studies, it is now part of Danaher Corporation with the SCIExe R&D division still located in Toronto, Canada.

















