Related Research Articles

In immunology, an antigen (Ag) is a molecule, moiety, foreign particulate matter, or an allergen, such as pollen, that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response.

Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.
Glycomics is the comprehensive study of glycomes, including genetic, physiologic, pathologic, and other aspects. Glycomics "is the systematic study of all glycan structures of a given cell type or organism" and is a subset of glycobiology. The term glycomics is derived from the chemical prefix for sweetness or a sugar, "glyco-", and was formed to follow the omics naming convention established by genomics and proteomics.

Glycoproteins are proteins which contain oligosaccharide (sugar) chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosylation. Secreted extracellular proteins are often glycosylated.
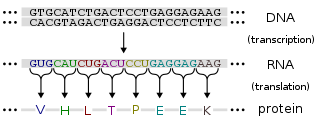
Protein production is the biotechnological process of generating a specific protein. It is typically achieved by the manipulation of gene expression in an organism such that it expresses large amounts of a recombinant gene. This includes the transcription of the recombinant DNA to messenger RNA (mRNA), the translation of mRNA into polypeptide chains, which are ultimately folded into functional proteins and may be targeted to specific subcellular or extracellular locations.
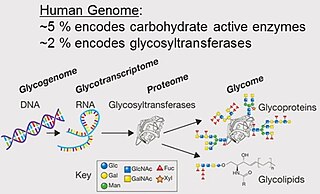
A glycome is the entire complement or complete set of all sugars, whether free or chemically bound in more complex molecules, of an organism. An alternative definition is the entirety of carbohydrates in a cell. The glycome may in fact be one of the most complex entities in nature. "Glycomics, analogous to genomics and proteomics, is the systematic study of all glycan structures of a given cell type or organism" and is a subset of glycobiology.

A monoclonal antibody is an antibody produced from a cell lineage made by cloning a unique white blood cell. All subsequent antibodies derived this way trace back to a unique parent cell.
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The part of an antibody that binds to the epitope is called a paratope. Although epitopes are usually non-self proteins, sequences derived from the host that can be recognized are also epitopes.

Chinese hamster ovary (CHO) cells are a family of immortalized cell lines derived from epithelial cells of the ovary of the Chinese hamster, often used in biological and medical research and commercially in the production of recombinant therapeutic proteins. They have found wide use in studies of genetics, toxicity screening, nutrition and gene expression, and particularly since the 1980s to express recombinant proteins. CHO cells are the most commonly used mammalian hosts for industrial production of recombinant protein therapeutics.
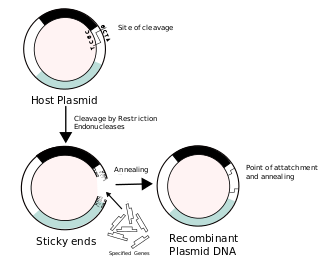
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
Pharming, a portmanteau of farming and pharmaceutical, refers to the use of genetic engineering to insert genes that code for useful pharmaceuticals into host animals or plants that would otherwise not express those genes, thus creating a genetically modified organism (GMO). Pharming is also known as molecular farming, molecular pharming, or biopharming.
A biopharmaceutical, also known as a biological medical product, or biologic, is any pharmaceutical drug product manufactured in, extracted from, or semisynthesized from biological sources. Different from totally synthesized pharmaceuticals, they include vaccines, whole blood, blood components, allergenics, somatic cells, gene therapies, tissues, recombinant therapeutic protein, and living medicines used in cell therapy. Biologics can be composed of sugars, proteins, nucleic acids, or complex combinations of these substances, or may be living cells or tissues. They are isolated from living sources—human, animal, plant, fungal, or microbial. They can be used in both human and animal medicine.
Immunogenicity is the ability of a foreign substance, such as an antigen, to provoke an immune response in the body of a human or other animal. It may be wanted or unwanted:
Humanized antibodies are antibodies from non-human species whose protein sequences have been modified to increase their similarity to antibody variants produced naturally in humans. The process of "humanization" is usually applied to monoclonal antibodies developed for administration to humans. Humanization can be necessary when the process of developing a specific antibody involves generation in a non-human immune system. The protein sequences of antibodies produced in this way are partially distinct from homologous antibodies occurring naturally in humans, and are therefore potentially immunogenic when administered to human patients. The International Nonproprietary Names of humanized antibodies end in -zumab, as in omalizumab.

Fusion proteins or chimeric (kī-ˈmir-ik) proteins are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this fusion gene results in a single or multiple polypeptides with functional properties derived from each of the original proteins. Recombinant fusion proteins are created artificially by recombinant DNA technology for use in biological research or therapeutics. Chimeric or chimera usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. Chimeric mutant proteins occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncoproteins. The bcr-abl fusion protein is a well-known example of an oncogenic fusion protein, and is considered to be the primary oncogenic driver of chronic myelogenous leukemia.
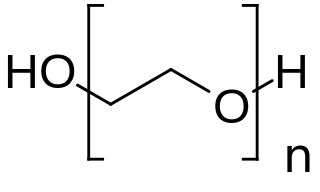
PEGylation is the process of both covalent and non-covalent attachment or amalgamation of polyethylene glycol polymer chains to molecules and macrostructures, such as a drug, therapeutic protein or vesicle, which is then described as PEGylated. PEGylation affects the resulting derivatives or aggregates interactions, which typically slows down their coalescence and degradation as well as elimination in vivo.
Biomolecular engineering is the application of engineering principles and practices to the purposeful manipulation of molecules of biological origin. Biomolecular engineers integrate knowledge of biological processes with the core knowledge of chemical engineering in order to focus on molecular level solutions to issues and problems in the life sciences related to the environment, agriculture, energy, industry, food production, biotechnology and medicine.
A subunit vaccine is a vaccine that contains purified parts of the pathogen that are antigenic, or necessary to elicit a protective immune response. Subunit vaccine can be made from dissembled viral particles in cell culture or recombinant DNA expression, in which case it is a recombinant subunit vaccine.
Mass spectrometric immunoassay (MSIA) is a rapid method is used to detect and/ or quantify antigens and or antibody analytes. This method uses an analyte affinity isolation to extract targeted molecules and internal standards from biological fluid in preparation for matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). This method allows for "top down" and "bottom up" analysis. This sensitive method allows for a new and improved process for detecting multiple antigens and antibodies in a single assay. This assay is also capable of distinguishing mass shifted forms of the same molecule via a panantibody, as well as distinguish point mutations in proteins. Each specific form is detected uniquely based on their characteristic molecular mass. MSIA has dual specificity because of the antibody-antigen reaction coupled with the power of a mass spectrometer.
Recombinant antibodies are antibody fragments produced by using recombinant antibody coding genes. They mostly consist of a heavy and light chain of the variable region of immunoglobulin. Recombinant antibodies have many advantages in both medical and research applications, which make them a popular subject of exploration and new production against specific targets. The most commonly used form is the single chain variable fragment (scFv), which has shown the most promising traits exploitable in human medicine and research. In contrast to monoclonal antibodies produced by hybridoma technology, which may lose the capacity to produce the desired antibody over time or the antibody may undergo unwanted changes, which affect its functionality, recombinant antibodies produced in phage display maintain high standard of specificity and low immunogenicity.
References
- 1 2 "Tracking Host Cell Proteins During Biopharmaceutical Manufacturing: Advanced Methodologies to Ensure High Product Quality". www.americanpharmaceuticalreview.com. Retrieved 2018-10-02.
- ↑ Goey CH, Alhuthali S, Kontoravdi C (2018). "Host cell protein removal from biopharmaceutical preparations: Towards the implementation of quality by design". Biotechnology Advances. 36 (4): 1223–1237. doi:10.1016/j.biotechadv.2018.03.021. hdl: 10044/1/85952 . PMID 29654903. S2CID 4870812.
- ↑ Dimitrov DS (2012). "Therapeutic Proteins". Methods in Molecular Biology. Vol. 899. pp. 1–26. doi:10.1007/978-1-61779-921-1_1. ISBN 978-1-61779-920-4. ISSN 1940-6029. PMC 6988726 . PMID 22735943.
- 1 2 3 Zhu-Shimoni J, Yu C, Nishihara J, Wong RM, Gunawan F, Lin M, et al. (December 2014). "Host cell protein testing by ELISAs and the use of orthogonal methods". Biotechnology and Bioengineering. 111 (12): 2367–2379. doi: 10.1002/bit.25327 . PMID 24995961. S2CID 23923786.
- 1 2 3 4 5 6 Wang X, Hunter AK, Mozier NM (June 2009). "Host cell proteins in biologics development: Identification, quantitation and risk assessment". Biotechnology and Bioengineering. 103 (3): 446–458. doi: 10.1002/bit.22304 . PMID 19388135. S2CID 22707536.
- 1 2 3 Bracewell DG, Francis R, Smales CM (September 2015). "The future of host cell protein (HCP) identification during process development and manufacturing linked to a risk-based management for their control". Biotechnology and Bioengineering. 112 (9): 1727–1737. doi:10.1002/bit.25628. PMC 4973824 . PMID 25998019.
- ↑ Guiochon G, Beaver LA (December 2011). "Separation science is the key to successful biopharmaceuticals". Journal of Chromatography A. 1218 (49): 8836–8858. doi:10.1016/j.chroma.2011.09.008. PMID 21982447.
- ↑ Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, et al. (September 1997). "The complete genome sequence of Escherichia coli K-12". Science. 277 (5331): 1453–1462. doi: 10.1126/science.277.5331.1453 . PMID 9278503.
- ↑ Zagulski M, Herbert CJ, Rytka J (1998). "Sequencing and functional analysis of the yeast genome". Acta Biochimica Polonica. 45 (3): 627–643. doi: 10.18388/abp.1998_4201 . PMID 9918489.
- ↑ Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. (December 2002). "Initial sequencing and comparative analysis of the mouse genome". Nature. 420 (6915): 520–562. Bibcode:2002Natur.420..520W. doi: 10.1038/nature01262 . PMID 12466850.
- ↑ Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. (April 2004). "Genome sequence of the Brown Norway rat yields insights into mammalian evolution". Nature. 428 (6982): 493–521. Bibcode:2004Natur.428..493G. doi: 10.1038/nature02426 . PMID 15057822.
- ↑ "The Sequence of the Human Genome". Science. 291 (5507): 1155.4–1155. 2001-02-16. doi:10.1126/science.291.5507.1155d. ISSN 0036-8075. S2CID 196263909.
- 1 2 Pilely K, Johansen MR, Lund RR, Kofoed T, Jørgensen TK, Skriver L, et al. (January 2022). "Monitoring process-related impurities in biologics-host cell protein analysis". Analytical and Bioanalytical Chemistry. 414 (2): 747–758. doi:10.1007/s00216-021-03648-2. PMC 8483941 . PMID 34595561.
- ↑ Heissel S, Bunkenborg J, Kristiansen MP, Holmbjerg AF, Grimstrup M, Mørtz E, et al. (July 2018). "Evaluation of spectral libraries and sample preparation for DIA-LC-MS analysis of host cell proteins: A case study of a bacterially expressed recombinant biopharmaceutical protein" (PDF). Protein Expression and Purification. 147: 69–77. doi:10.1016/j.pep.2018.03.002. PMID 29526817. S2CID 4741772.