
Acidobacteriota is a phylum of Gram-negative bacteria. Its members are physiologically diverse and ubiquitous, especially in soils, but are under-represented in culture.
Methylotrophs are a diverse group of microorganisms that can use reduced one-carbon compounds, such as methanol or methane, as the carbon source for their growth; and multi-carbon compounds that contain no carbon-carbon bonds, such as dimethyl ether and dimethylamine. This group of microorganisms also includes those capable of assimilating reduced one-carbon compounds by way of carbon dioxide using the ribulose bisphosphate pathway. These organisms should not be confused with methanogens which on the contrary produce methane as a by-product from various one-carbon compounds such as carbon dioxide. Some methylotrophs can degrade the greenhouse gas methane, and in this case they are called methanotrophs. The abundance, purity, and low price of methanol compared to commonly used sugars make methylotrophs competent organisms for production of amino acids, vitamins, recombinant proteins, single-cell proteins, co-enzymes and cytochromes.
Nitrospira translate into "a nitrate spiral" is a genus of bacteria within the monophyletic clade of the Nitrospirota phylum. The first member of this genus was described 1986 by Watson et al., isolated from the Gulf of Maine. The bacterium was named Nitrospira marina. Populations were initially thought to be limited to marine ecosystems, but it was later discovered to be well-suited for numerous habitats, including activated sludge of wastewater treatment systems, natural biological marine settings, water circulation biofilters in aquarium tanks, terrestrial systems, fresh and salt water ecosystems, agricultural lands and hot springs. Nitrospira is a ubiquitous bacterium that plays a role in the nitrogen cycle by performing nitrite oxidation in the second step of nitrification. Nitrospira live in a wide array of environments including but not limited to, drinking water systems, waste treatment plants, rice paddies, forest soils, geothermal springs, and sponge tissue. Despite being abundant in many natural and engineered ecosystems Nitrospira are difficult to culture, so most knowledge of them is from molecular and genomic data. However, due to their difficulty to be cultivated in laboratory settings, the entire genome was only sequenced in one species, Nitrospira defluvii. In addition, Nitrospira bacteria's 16S rRNA sequences are too dissimilar to use for PCR primers, thus some members go unnoticed. In addition, members of Nitrospira with the capabilities to perform complete nitrification has also been discovered and cultivated.
Dehalococcoides is a genus of bacteria within class Dehalococcoidia that obtain energy via the oxidation of hydrogen and subsequent reductive dehalogenation of halogenated organic compounds in a mode of anaerobic respiration called organohalide respiration. They are well known for their great potential to remediate halogenated ethenes and aromatics. They are the only bacteria known to transform highly chlorinated dioxins, PCBs. In addition, they are the only known bacteria to transform tetrachloroethene to ethene.
Methylorubrum extorquens is a Gram-negative bacterium. Methylorubrum species often appear pink, and are classified as pink-pigmented facultative methylotrophs, or PPFMs. The wild type has been known to use both methane and multiple carbon compounds as energy sources. Specifically, M. extorquens has been observed to use primarily methanol and C1 compounds as substrates in their energy cycles. It has been also observed that use lanthanides as a cofactor to increase its methanol dehydrogenase activity
In taxonomy, Pseudovibrio is a genus of the Hyphomicrobiales. Bacteria belonging to this genus have been often isolated from marine invertebrates and have been described to be metabolically versatile. Recent comparative genomic analyses revealed that these organisms have the genomic potential to produce a great array of systems to interact with their hosts, including type III, IV, VI secretion systems and different type of toxin-like proteins. Moreover, in their genomes several biosynthetic gene clusters producing potentially novel bioactive compounds were recently identified.
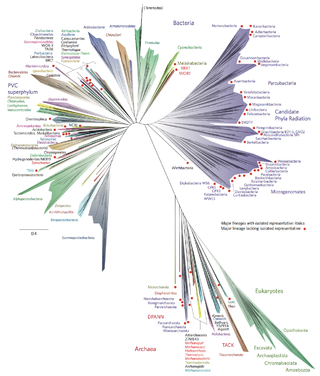
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.

2,4-Diacetylphloroglucinol or Phl is a natural phenol found in several bacteria:
Cycloclasticus is a genus in the phylum Pseudomonadota (Bacteria).
Methylocella silvestris is a bacterium from the genus Methylocella spp which are found in many acidic soils and wetlands. Historically, Methylocella silvestris was originally isolated from acidic forest soils in Germany, and it is described as Gram-negative, aerobic, non-pigmented, non-motile, rod-shaped and methane-oxidizing facultative methanotroph. As an aerobic methanotrophic bacteria, Methylocella spp use methane (CH4), and methanol as their main carbon and energy source, as well as multi compounds acetate, pyruvate, succinate, malate, and ethanol. They were known to survive in the cold temperature from 4° to 30° degree of Celsius with the optimum at around 15° to 25 °C, but no more than 36 °C. They grow better in the pH scale between 4.5 to 7.0. It lacks intracytoplasmic membranes common to all methane-oxidizing bacteria except Methylocella, but contain a vesicular membrane system connected to the cytoplasmic membrane. BL2T (=DSM 15510T=NCIMB 13906T) is the type strain.
Desulfitobacterium dehalogenans is a species of bacteria. They are facultative organohalide respiring bacteria capable of reductively dechlorinating chlorophenolic compounds and tetrachloroethene. They are anaerobic, motile, Gram-positive and rod-shaped bacteria capable of utilizing a wide range of electron donors and acceptors. The type strain JW/IU-DCT, DSM 9161, NCBi taxonomy ID 756499.
Desulfitobacterium hafniense is a species of gram positive bacteria, its type strain is DCB-2T..
Pelobacter carbinolicus is a species of bacteria that ferments 2,3-butanediol and acetoin. It is Gram-negative, strictly anaerobic and non-spore-forming. Gra Bd 1 is the type strain. Its genome has been sequenced.
Smithella propionica is a species of bacteria, the type species of its genus. It is anaerobic, syntrophic, propionate-oxidizing bacteria, with type strain LYPT.
Desulfitobacterium metallireducens is an anaerobic bacterium that couples growth to the reduction of metals and humic acids as well as chlorinated compounds. Its type strain is 853-15A(T). It was first isolated from a uranium-contaminated aquifer sediment.
Treponema primitia is a bacterium, the first termite gut spirochete to be isolated, together with Treponema azotonutricium.
Roseivivax halodurans is a species of bacteria, the type species of its genus. It is aerobic and bacteriochlorophyll-containing, first isolated from the charophytes on the stromatolites of a saline lake located on the west coast of Australia. It is chemoheterotrophic, Gram-negative, motile, rod-shaped and with subpolar flagella. Its type strain is OCh 239T.
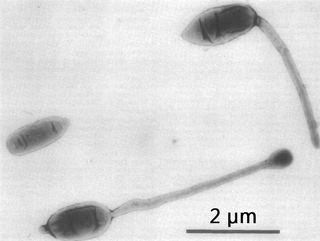
Hyphomicrobium is a genus of Gram-negative, non-spore-forming, rod-shaped bacteria from the family of Hyphomicrobiaceae. It has a large polar or sub-polar filiform prostheca very similar to that of Caulobacter. In addition to having a nutritional function, the prostheca also plays a role in the initiation of DNA replication.
Hyphomicrobium sulfonivorans is a bacterium from the genus of Hyphomicrobium which was isolated from garden soil in Warwickshire in England.
Methylophaga thiooxydans is a methylotrophic bacterium that requires high salt concentrations for growth. It was originally isolated from a culture of the algae Emiliania huxleyi, where it grows by breaking down dimethylsulfoniopropionate from E. hexleyi into dimethylsulfide and acrylate. M. thiooxydans has been implicated as a dominant organism in phytoplankton blooms, where it consumes dimethylsulfide, methanol and methyl bromide released by dying phytoplankton. It was also identified as one of the dominant organisms present in the plume following the Deepwater Horizon oil spill, and was identified as a major player in the breakdown of methanol in coastal surface water in the English Channel.



