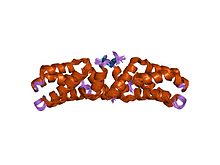
Intermediate filaments (IFs) are cytoskeletal structural components found in the cells of vertebrates, and many invertebrates. Homologues of the IF protein have been noted in an invertebrate, the cephalochordate Branchiostoma.
A coiled coil is a structural motif in proteins in which 2–7 alpha-helices are coiled together like the strands of a rope. They have been found in roughly 5-10% of proteins and have a variety of functions. They are one of the most widespread motifs found in protein-protein interactions. To aid protein study, several tools have been developed to predict coiled-coils in protein structures. Many coiled coil-type proteins are involved in important biological functions, such as the regulation of gene expression — e.g., transcription factors. Notable examples are the oncoproteins c-Fos and c-Jun, as well as the muscle protein tropomyosin.
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits, and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits, or two heterodimer subunits.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.
A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

Signal recognition particle (SRP) receptor, also called the docking protein, is a dimer composed of 2 different subunits that are associated exclusively with the rough ER in mammalian cells. Its main function is to identify the SRP units. SRP is a molecule that helps the ribosome-mRNA-polypeptide complexes to settle down on the membrane of the endoplasmic reticulum.

The breakpoint cluster region protein (BCR) also known as renal carcinoma antigen NY-REN-26 is a protein that in humans is encoded by the BCR gene. BCR is one of the two genes in the BCR-ABL fusion protein, which is associated with the Philadelphia chromosome. Two transcript variants encoding different isoforms have been found for this gene.

Transforming growth factor, beta receptor II (70/80kDa) is a TGF beta receptor. TGFBR2 is its human gene.

MAGUK p55 subfamily member 5 is a protein that in humans is encoded by the MPP5 gene. Members of the peripheral membrane-associated guanylate kinase (MAGUK) family function in tumor suppression and receptor clustering by forming multiprotein complexes containing distinct sets of transmembrane, cytoskeletal, and cytoplasmic signaling proteins. All MAGUKs contain a PDZ-SH3-GUK core and are divided into 4 subfamilies, DLG-like, ZO1-like, p55-like, and LIN2-like, based on their size and the presence of additional domains. MPP5 is a member of the p55-like MAGUK subfamily.[supplied by OMIM]

Dishevelled (Dsh) is a family of proteins involved in canonical and non-canonical Wnt signalling pathways. Dsh is a cytoplasmic phosphoprotein that acts directly downstream of frizzled receptors. It takes its name from its initial discovery in flies, where a mutation in the dishevelled gene was observed to cause improper orientation of body and wing hairs. There are vertebrate homologs in zebrafish, Xenopus (Xdsh), mice and humans. Dsh relays complex Wnt signals in tissues and cells, in normal and abnormal contexts. It is thought to interact with the SPATS1 protein when regulating the Wnt Signalling pathway.

E1 is one of two subunits of the envelope glycoprotein found in the hepatitis C virus. The other subunit is E2. This protein is a type 1 transmembrane protein with a highly glycosylated N-terminal ectodomain and a C-terminal hydrophobic anchor. After being synthesized the E1 glycoproteins associates with the E2 glycoprotein as a noncovalent heterodimer.
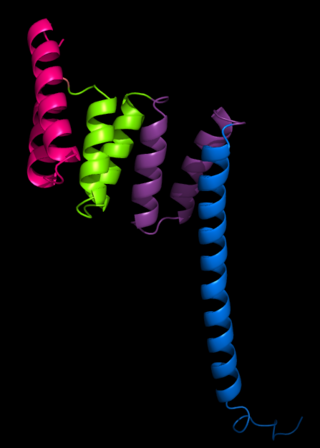
The tetratricopeptide repeat (TPR) is a structural motif. It consists of a degenerate 34 amino acid tandem repeat identified in a wide variety of proteins. It is found in tandem arrays of 3–16 motifs, which form scaffolds to mediate protein–protein interactions and often the assembly of multiprotein complexes. These alpha-helix pair repeats usually fold together to produce a single, linear solenoid domain called a TPR domain. Proteins with such domains include the anaphase-promoting complex (APC) subunits cdc16, cdc23 and cdc27, the NADPH oxidase subunit p67-phox, hsp90-binding immunophilins, transcription factors, the protein kinase R (PKR), the major receptor for peroxisomal matrix protein import PEX5, protein arginine methyltransferase 9 (PRMT9), and mitochondrial import proteins.
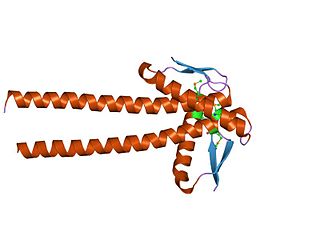
In molecular biology, the ZapA protein family is a group of related proteins that includes the cell division protein ZapA. The structure of ZapA has a core structure consisting of two layers alpha/beta, and has a long C-terminal helix that forms dimeric parallel and tetrameric antiparallel coiled coils. ZapA interacts with FtsZ, where FtsZ is part of a mid-cell cytokinetic structure termed the Z-ring that recruits a hierarchy of fission related proteins early in the bacterial cell cycle. ZapA drives the polymerisation and filament bundling of FtsZ, thereby contributing to the spatio-temporal tuning of the Z-ring.
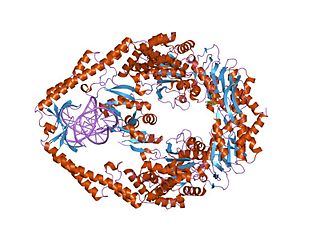
MutS is a mismatch DNA repair protein, originally described in Escherichia coli.

In molecular biology, the ARID domain ) is a protein domain that binds to DNA. ARID domain-containing proteins are found in fungi, plants and invertebrate and vertebrate metazoans. ARID-encoding genes are involved in a variety of biological processes including embryonic development, cell lineage gene regulation and cell cycle control. Although the specific roles of this domain and of ARID-containing proteins in transcriptional regulation are yet to be elucidated, they include both positive and negative transcriptional regulation and a likely involvement in the modification of chromatin structure. The basic structure of the ARID domain appears to be a series of six alpha-helices separated by beta-strands, loops, or turns, but the structured region may extend to an additional helix at either or both ends of the basic six. Based on primary sequence homology, they can be partitioned into three structural classes: Minimal ARID proteins that consist of a core domain formed by six alpha helices; ARID proteins that supplement the core domain with an N-terminal alpha-helix; and Extended-ARID proteins, which contain the core domain and additional alpha-helices at their N- and C-termini.
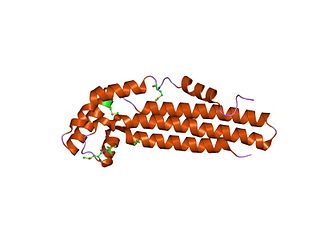
In molecular biology, CagZ is a protein produced by Helicobacter pylori. It is a 23 kDa protein consisting of a single compact L-shaped domain, composed of seven alpha-helices that run antiparallel to each other. 70% of the amino acids are in alpha-helix conformation and no beta-sheet is present. CagZ is essential for the translocation of the pathogenic protein CagA into host cells.
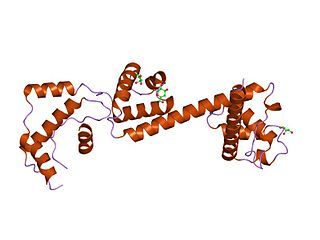
In molecular biology, the HAND domain is a protein domain which adopts a secondary structure consisting of four alpha helices, three of which form an L-like configuration. Helix H2 runs antiparallel to helices H3 and H4, packing closely against helix H4, whilst helix H1 reposes in the concave surface formed by these three helices and runs perpendicular to them. This domain confers DNA and nucleosome binding properties to the proteins in which it occurs. It is named the HAND domain because its 4-helical structure resembles an open hand.

In molecular biology, the IMD domain is a BAR-like domain of approximately 250 amino acids found at the N-terminus in the insulin receptor tyrosine kinase substrate p53 (IRSp53/BAIAP2) and in the evolutionarily related IRSp53/MIM (MTSS1) family. In IRSp53, a ubiquitous regulator of the actin cytoskeleton, the IMD domain acts as conserved F-actin bundling domain involved in filopodium formation. Filopodium-inducing IMD activity is regulated by Cdc42 and Rac1 and is SH3-independent. The IRSp53/MIM family is a novel F-actin bundling protein family that includes invertebrate relatives:

The C-terminal domain ofBeta2-adaptin is a protein domain is involved in cell trafficking by aiding import and export of substances in and out of the cell.