
Chlamydomonas reinhardtii is a single-cell green alga about 10 micrometres in diameter that swims with two flagella. It has a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an eyespot that senses light.

Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more mass analyzers are coupled together using an additional reaction step to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.

Peptide mass fingerprinting (PMF) is an analytical technique for protein identification in which the unknown protein of interest is first cleaved into smaller peptides, whose absolute masses can be accurately measured with a mass spectrometer such as MALDI-TOF or ESI-TOF. The method was developed in 1993 by several groups independently. The peptide masses are compared to either a database containing known protein sequences or even the genome. This is achieved by using computer programs that translate the known genome of the organism into proteins, then theoretically cut the proteins into peptides, and calculate the absolute masses of the peptides from each protein. They then compare the masses of the peptides of the unknown protein to the theoretical peptide masses of each protein encoded in the genome. The results are statistically analyzed to find the best match.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.
A tandem mass tag (TMT) is a chemical label that facilitates sample multiplexing in mass spectrometry (MS)-based quantification and identification of biological macromolecules such as proteins, peptides and nucleic acids. TMT belongs to a family of reagents referred to as isobaric mass tags which are a set of molecules with the same mass, but yield reporter ions of differing mass after fragmentation. The relative ratio of the measured reporter ions represents the relative abundance of the tagged molecule, although ion suppression has a detrimental effect on accuracy. Despite these complications, TMT-based proteomics has been shown to afford higher precision than Label-free quantification. In addition to aiding in protein quantification, TMT tags can also increase the detection sensitivity of certain highly hydrophilic analytes, such as phosphopeptides, in RPLC-MS analyses.
Shotgun proteomics refers to the use of bottom-up proteomics techniques in identifying proteins in complex mixtures using a combination of high performance liquid chromatography combined with mass spectrometry. The name is derived from shotgun sequencing of DNA which is itself named after the rapidly expanding, quasi-random firing pattern of a shotgun. The most common method of shotgun proteomics starts with the proteins in the mixture being digested and the resulting peptides are separated by liquid chromatography. Tandem mass spectrometry is then used to identify the peptides.
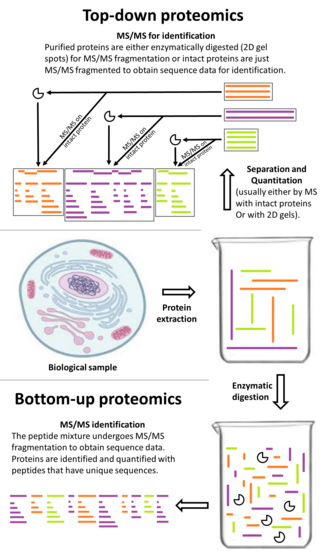
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE), preparative one-dimensional gel electrophoresis, or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.
Michael L. Gross is Professor of Chemistry, Medicine, and Immunology, at Washington University in St. Louis. He was formerly Professor of Chemistry at the University of Nebraska-Lincoln from 1968–1994. He is recognized for his contributions to the field of mass spectrometry and ion chemistry. He is credited with the discovery of distonic ions, chemical species containing a radical and an ionic site on different atoms of the same molecule.
Label-free quantification is a method in mass spectrometry that aims to determine the relative amount of proteins in two or more biological samples. Unlike other methods for protein quantification, label-free quantification does not use a stable isotope containing compound to chemically bind to and thus label the protein.

Capillary electrophoresis–mass spectrometry (CE–MS) is an analytical chemistry technique formed by the combination of the liquid separation process of capillary electrophoresis with mass spectrometry. CE–MS combines advantages of both CE and MS to provide high separation efficiency and molecular mass information in a single analysis. It has high resolving power and sensitivity, requires minimal volume and can analyze at high speed. Ions are typically formed by electrospray ionization, but they can also be formed by matrix-assisted laser desorption/ionization or other ionization techniques. It has applications in basic research in proteomics and quantitative analysis of biomolecules as well as in clinical medicine. Since its introduction in 1987, new developments and applications have made CE-MS a powerful separation and identification technique. Use of CE–MS has increased for protein and peptides analysis and other biomolecules. However, the development of online CE–MS is not without challenges. Understanding of CE, the interface setup, ionization technique and mass detection system is important to tackle problems while coupling capillary electrophoresis to mass spectrometry.

Isobaric labeling is a mass spectrometry strategy used in quantitative proteomics. Peptides or proteins are labeled with chemical groups that have identical mass (isobaric), but vary in terms of distribution of heavy isotopes in their structure. These tags, commonly referred to as tandem mass tags, are designed so that the mass tag is cleaved at a specific linker region upon high-energy CID (HCD) during tandem mass spectrometry yielding reporter ions of different masses. The most common isobaric tags are amine-reactive tags. However, tags that react with cysteine residues and carbonyl groups have also been described. These amine-reactive groups go through N-hydroxysuccinimide (NHS) reactions, which are based around three types of functional groups. Isobaric labeling methods include tandem mass tags (TMT), isobaric tags for relative and absolute quantification (iTRAQ), mass differential tags for absolute and relative quantification, and dimethyl labeling. TMTs and iTRAQ methods are most common and developed of these methods. Tandem mass tags have a mass reporter region, a cleavable linker region, a mass normalization region, and a protein reactive group and have the same total mass.
Joshua Coon is a professor of chemistry and biomolecular chemistry and the inaugural holder of the Thomas and Margaret Pyle Chair at the University of Wisconsin–Madison, and an affiliate of the Morgridge Institute for Research.
Kevin Downard is a British - Australian academic scientist whose research specialises in the improving responses to infectious disease through the application and development of mass spectrometry and other molecular approaches in the life and medical sciences. Downard has over 35 years of experience in the field and has written over 145 lead-author scientific peer-reviewed journal publications, and two books including a textbook for the Royal Society of Chemistry and the first book to be published on the role of mass spectrometry in the study of protein interactions.

Albert J.R. Heck is a Dutch scientist and professor at Utrecht University, the Netherlands in the field of mass spectrometry and proteomics. He is known for his work on technologies to study proteins in their natural environment, with the aim to understand their biological function. Albert Heck was awarded the Spinoza Prize in 2017, the highest scientific award in the Netherlands.
Renã A. S. Robinson is an associate professor and the Dorothy J. Wingfield Phillips Chancellor's Faculty Fellow in the department of chemistry at the Vanderbilt University, where she is the principal investigator of the RASR Laboratory.
Skyline is an open source software for targeted proteomics and metabolomics data analysis. It runs on Microsoft Windows and supports the raw data formats from multiple mass spectrometric vendors. It contains a graphical user interface to display chromatographic data for individual peptide or small molecule analytes.

Mandë Holford is an associate professor in chemistry at Hunter College with scientific appointments at the American Museum of Natural History and Weill Cornell Medical College. Her interdisciplinary research covering 'mollusks to medicine' spans chemistry and biology and aims to discover, characterize, and deliver novel peptides from venomous marine snails as tools for manipulating cellular physiology in pain and cancer.
Sabeeha Sabanali Merchant is a professor of plant biology at the University of California, Berkeley. She studies the photosynthetic metabolism and metalloenzymes In 2010 Merchant led the team that sequenced the Chlamydomonas genome. She was elected a member of the National Academy of Sciences in 2012.










