
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and those that have one strand of DNA in their genome, called single-stranded DNA (ssDNA) viruses. dsDNA viruses primarily belong to two realms: Duplodnaviria and Varidnaviria, and ssDNA viruses are almost exclusively assigned to the realm Monodnaviria, which also includes some dsDNA viruses. Additionally, many DNA viruses are unassigned to higher taxa. Reverse transcribing viruses, which have a DNA genome that is replicated through an RNA intermediate by a reverse transcriptase, are classified into the kingdom Pararnavirae in the realm Riboviria.

The mung bean, alternatively known as green gram, mungo bean or mongo bean, is a plant species in the legume family. The mung bean is mainly cultivated in East, Southeast and South Asia. It is used as an ingredient in both savoury and sweet dishes.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.

Begomovirus is a genus of viruses, in the family Geminiviridae. They are plant viruses that as a group have a very wide host range, infecting dicotyledonous plants. Worldwide they are responsible for a considerable amount of economic damage to many important crops such as tomatoes, beans, squash, cassava and cotton. There are 445 species in this genus.
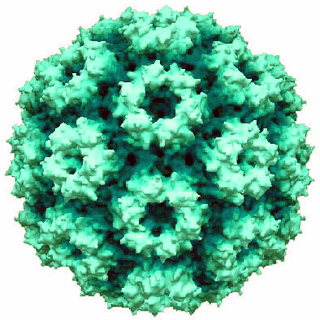
Cowpea chlorotic mottle virus, known by the abbreviation CCMV, is a virus that specifically infects the cowpea plant, or black-eyed pea. The leaves of infected plants develop yellow spots, hence the name "chlorotic". Similar to its "brother" virus, Cowpea mosaic virus (CPMV), CCMV is produced in high yield in plants. In the natural host, viral particles can be produced at 1–2 mg per gram of infected leaf tissue. Belonging to the bromovirus genus, cowpea chlorotic mottle virus (CCMV) is a small spherical plant virus. Other members of this genus include the brome mosaic virus (BMV) and the broad bean mottle virus (BBMV).

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.

Cassava mosaic virus is the common name used to refer to any of eleven different species of plant pathogenic virus in the genus Begomovirus. African cassava mosaic virus (ACMV), East African cassava mosaic virus (EACMV), and South African cassava mosaic virus (SACMV) are distinct species of circular single-stranded DNA viruses which are transmitted by whiteflies and primarily infect cassava plants; these have thus far only been reported from Africa. Related species of viruses are found in India and neighbouring islands, though cassava is cultivated in Latin America as well as Southeast Asia. Nine species of cassava-infecting geminiviruses have been identified between Africa and India based on genomic sequencing and phylogenetic analysis. This number is likely to grow due to a high rate of natural transformation associated with CMV.
Bean yellow mosaic virus is a plant pathogenic virus in the genus Potyvirus and the virus family Potyviridae. Like other members of the Potyvirus genus, it is a monopartite strand of positive-sense, single-stranded RNA surrounded by a capsid made for a single viral encoded protein. The virus is a filamentous particle that measures about 750 nm in length. This virus is transmitted by species of aphids and by mechanical inoculation.

Beet curly top virus (BCTV) is a pathogenic plant virus of the family Geminiviridae, containing a single-stranded DNA. The family Geminiviridae consists of nine genera based on their host range, virus genome structure, and type of insect vector. BCTV is a Curtovirus affecting hundreds of plants. The only known vector is the beet leafhopper, which is native to the Western United States.
Clover yellow mosaic virus (ClYMV) is a plant pathogenic virus in the genus Potexvirus and the virus family Alphaflexiviridae. Its flexuous rod-shaped particles measure about 539 nm in length.
Cotton leaf curl viruses (CLCuV) are a number of plant pathogenic virus species of the family Geminiviridae.
Tomato yellow leaf curl virus (TYLCV) is a DNA virus from the genus Begomovirus and the family Geminiviridae. TYLCV causes the most destructive disease of tomato, and it can be found in tropical and subtropical regions causing severe economic losses. This virus is transmitted by an insect vector from the family Aleyrodidae and order Hemiptera, the whitefly Bemisia tabaci, commonly known as the silverleaf whitefly or the sweet potato whitefly. The primary host for TYLCV is the tomato plant, and other plant hosts where TYLCV infection has been found include eggplants, potatoes, tobacco, beans, and peppers. Due to the rapid spread of TYLCV in the last few decades, there is an increased focus in research trying to understand and control this damaging pathogen. Some interesting findings include the virus being sexually transmitted from infected males to non-infected females, and an evidence that TYLCV is transovarially transmitted to offspring for two generations.
Alphasatellites are a single-stranded DNA family of satellite viruses that depend on the presence of another virus to replicate their genomes. As such, they have minimal genomes with very low genomic redundancy. The genome is a single circular single strand DNA molecule. The first alphasatellites were described in 1999 and were associated with cotton leaf curl disease and Ageratum yellow vein disease. As begomoviruses are being characterised at the molecular level an increasing number of alphasatellites are being described.

Watermelon mosaic virus (WMV) also known as Marrow mosaic virus, Melon mosaic virus, and until recently Watermelon mosaic virus type 2 (WMV-2), is a plant pathogenic virus that causes viral infection in many different plants. The virus itself is referred to as Watermelon Mosaic Virus II or WMV-2 and is an isolate of the U.S. WMV-2 is a ssRNA positive strand virus that is part of the Potyviridae or Potyvirus clade. Like all RNA viruses, it contains a protein capsid which protects the inner viral RNA. First described on squash in Florida, WMV arose from a unique recombination of genetic material contributed by Soybean mosaic virus (SMV) and Bean common mosaic virus (BCMV) along with Peanut Stripe virus (PSV).
Mastrevirus is a genus of ssDNA viruses, in the family Geminiviridae. Mostly monocotyledonous plants serve as natural hosts. They are vectored by planthoppers. There are 45 species in this genus. Diseases associated with this genus include: maize streak virus: maize streak disease (MSD).

Nanovirus is a genus of viruses, in the family Nanoviridae. Legume plants serve as natural hosts. There are 11 species in this genus. Diseases associated with this genus include: stunting, severe necrosis and early plant death.

Pepper leaf curl virus(PepLCV) is a DNA virus from the genus Begomovirus and the family Geminiviridae. PepLCV causes severe disease especially in pepper (Capsicum spp.). It can be found in tropical and subtropical regions such as Thailand and India, but has also been detected in countries such as the United States and Nigeria. This virus is transmitted by an insect vector from the family Aleyrodidae and order Hemiptera, the whitefly Bemisia tabaci. The primary host for PepLCV are several Capsicum spp.. PepLCV has been responsible for several epidemics and causes severe economic losses. It is the focus of research trying to understand the genetic basis of resistance. Currently, a source of resistance to the virus has been identified in the Bhut Jolokia pepper.
Papaya leaf curl virus(PaLCuV) is a DNA virus from the genus Begomovirus and the family Geminiviridae. PaLCuV causes severe disease in papaya (Carica papaya), but can sometimes infect other crops such as tobacco or tomato. It can be found in tropical and subtropical regions primarily in India, but closely related species have also been detected in countries such as China, Malaysia, Nigeria and South Korea. This virus is transmitted by an insect vector from the family Aleyrodidae and order Hemiptera, the whitefly Bemisia tabaci. PaLCuV has been responsible for several epidemics and causes severe economic losses. Because of the broad diversity of these viruses, their characterization and control remains difficult.
Sweet potato leaf curl virus is commonly abbreviated SPLCV. Select isolates are referred to as SPLCV followed by an abbreviation of where they were isolated. For example, the Brazilian isolate is referred to as SPLCV-Br.

Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.
Navneet Kumar Yadav, Central Drug Research Institute(CDRI), Lucknow INDIA .












