
Whiteflies are Hemipterans that typically feed on the undersides of plant leaves. They comprise the family Aleyrodidae, the only family in the superfamily Aleyrodoidea. More than 1550 species have been described.

Tobamovirus is a genus of positive-strand RNA viruses in the family Virgaviridae. Many plants, including tobacco, potato, tomato, and squash, serve as natural hosts. Diseases associated with this genus include: necrotic lesions on leaves. The name Tobamovirus comes from the host and symptoms of the first virus discovered.

Potyvirus is a genus of positive-strand RNA viruses in the family Potyviridae. Plants serve as natural hosts. Like begomoviruses, members of this genus may cause significant losses in agricultural, pastoral, horticultural, and ornamental crops. More than 200 species of aphids spread potyviruses, and most are from the subfamily Aphidinae. The genus contains 190 species and potyviruses account for about thirty percent of all currently known plant viruses.
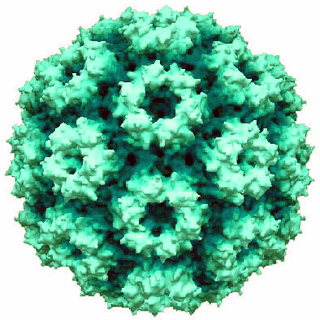
Cowpea chlorotic mottle virus, known by the abbreviation CCMV, is a virus that specifically infects the cowpea plant, or black-eyed pea. The leaves of infected plants develop yellow spots, hence the name "chlorotic". Similar to its "brother" virus, Cowpea mosaic virus (CPMV), CCMV is produced in high yield in plants. In the natural host, viral particles can be produced at 1–2 mg per gram of infected leaf tissue. Belonging to the bromovirus genus, cowpea chlorotic mottle virus (CCMV) is a small spherical plant virus. Other members of this genus include the brome mosaic virus (BMV) and the broad bean mottle virus (BBMV).
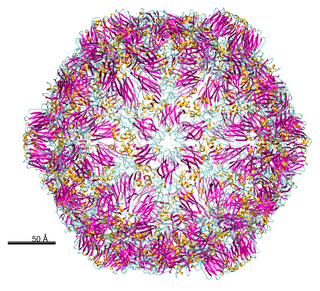
Nepovirus is a genus of viruses in the order Picornavirales, in the family Secoviridae, in the subfamily Comovirinae. Plants serve as natural hosts. There are 40 species in this genus. Nepoviruses, unlike the other two genera in the subfamily Comovirinae, are transmitted by nematodes.

Cassava mosaic virus is the common name used to refer to any of eleven different species of plant pathogenic virus in the genus Begomovirus. African cassava mosaic virus (ACMV), East African cassava mosaic virus (EACMV), and South African cassava mosaic virus (SACMV) are distinct species of circular single-stranded DNA viruses which are transmitted by whiteflies and primarily infect cassava plants; these have thus far only been reported from Africa. Related species of viruses are found in India and neighbouring islands, though cassava is cultivated in Latin America as well as Southeast Asia. Nine species of cassava-infecting geminiviruses have been identified between Africa and India based on genomic sequencing and phylogenetic analysis. This number is likely to grow due to a high rate of natural transformation associated with CMV.
Cotton leaf curl viruses (CLCuV) are a number of plant pathogenic virus species of the family Geminiviridae.
Tomato yellow leaf curl virus (TYLCV) is a DNA virus from the genus Begomovirus and the family Geminiviridae. TYLCV causes the most destructive disease of tomato, and it can be found in tropical and subtropical regions causing severe economic losses. This virus is transmitted by an insect vector from the family Aleyrodidae and order Hemiptera, the whitefly Bemisia tabaci, commonly known as the silverleaf whitefly or the sweet potato whitefly. The primary host for TYLCV is the tomato plant, and other plant hosts where TYLCV infection has been found include eggplants, potatoes, tobacco, beans, and peppers. Due to the rapid spread of TYLCV in the last few decades, there is an increased focus in research trying to understand and control this damaging pathogen. Some interesting findings include the virus being sexually transmitted from infected males to non-infected females, and an evidence that TYLCV is transovarially transmitted to offspring for two generations.
Alphasatellites are a single-stranded DNA family of satellite viruses that depend on the presence of another virus to replicate their genomes. As such, they have minimal genomes with very low genomic redundancy. The genome is a single circular single strand DNA molecule. The first alphasatellites were described in 1999 and were associated with cotton leaf curl disease and Ageratum yellow vein disease. As begomoviruses are being characterised at the molecular level an increasing number of alphasatellites are being described.
Ipomovirus is a genus of positive-strand RNA viruses in the family Potyviridae. Member viruses infect plants and are transmitted by whiteflies. The name of the genus is derived from Ipomoea – the generic name of sweet potato. There are seven species in this genus.
Bean calico mosaic virus is a plant virus transmitted by whiteflies that infects bean genera and species within the families Fabaceae, Malvaceae, and Solanaceae. Like other New World begomoviruses, its genome is bipartite, or having two parts. Phylogenetic analysis of its two genome segments, DNA-A and DNA-B, indicate the virus is from Sonora, Mexico, and shares a most recent common ancestor with the Leaf curl virus-E strain and the Texas pepper virus, both also found in the Sonora desert, and the Cabbage leaf curl virus from Florida.
Badnavirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There are 67 species in this genus. Diseases associated with this genus include: CSSV: leaf chlorosis, root necrosis, red vein banding in young leaves, small mottled pods, and stem/root swelling followed by die-back. Infection decreases yield by 25% within one year, 50% within two years and usually kills trees within 3–4 years.
Mastrevirus is a genus of ssDNA viruses, in the family Geminiviridae. Mostly monocotyledonous plants serve as natural hosts. They are vectored by planthoppers. There are 45 species in this genus. Diseases associated with this genus include: maize streak virus: maize streak disease (MSD).

Pepper leaf curl virus(PepLCV) is a DNA virus from the genus Begomovirus and the family Geminiviridae. PepLCV causes severe disease especially in pepper (Capsicum spp.). It can be found in tropical and subtropical regions such as Thailand and India, but has also been detected in countries such as the United States and Nigeria. This virus is transmitted by an insect vector from the family Aleyrodidae and order Hemiptera, the whitefly Bemisia tabaci. The primary host for PepLCV are several Capsicum spp.. PepLCV has been responsible for several epidemics and causes severe economic losses. It is the focus of research trying to understand the genetic basis of resistance. Currently, a source of resistance to the virus has been identified in the Bhut Jolokia pepper.
Papaya leaf curl virus(PaLCuV) is a DNA virus from the genus Begomovirus and the family Geminiviridae. PaLCuV causes severe disease in papaya (Carica papaya), but can sometimes infect other crops such as tobacco or tomato. It can be found in tropical and subtropical regions primarily in India, but closely related species have also been detected in countries such as China, Malaysia, Nigeria and South Korea. This virus is transmitted by an insect vector from the family Aleyrodidae and order Hemiptera, the whitefly Bemisia tabaci. PaLCuV has been responsible for several epidemics and causes severe economic losses. Because of the broad diversity of these viruses, their characterization and control remains difficult.
Sweet potato leaf curl virus is commonly abbreviated SPLCV. Select isolates are referred to as SPLCV followed by an abbreviation of where they were isolated. For example, the Brazilian isolate is referred to as SPLCV-Br.
Elijah Miinda Ateka is a Professor of Plant Virology at the Jomo Kenyatta University of Agriculture and Technology. He is involved with the diagnosis and characterisation of the sweet potato virus and the cassava virus, and is part of the Cassava Virus Action Project (CVAP).
Tomato yellow leaf curl China virus (TYLCCNV) is a virus which contains 25 isolates. It infects plants as different as tobacco and tomato, as well as genetically modified plants. Petunias can be infected, but show no symptoms. The microbiology of the virus has been studied in the Chinese province of Yunnan. Tomato yellow leaf curl China virus belongs to the genus Begomovirus, which also contains the tomato leaf curl China virus.
Tolecusatellitidae is a incertae sedis ssDNA/ssDNA(+) family of biological satellites. The family contains two genera and 131 species. This family of viruses depend on the presence of another virus to replicate their genomes, as such they have minimal genomes with very low genomic redundancy.










