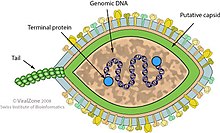
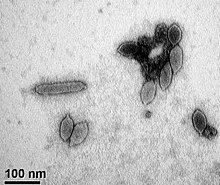
Virus classification is the process of naming viruses and placing them into a taxonomic system similar to the classification systems used for cellular organisms.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.

Φ6 is the best-studied bacteriophage of the virus family Cystoviridae. It infects Pseudomonas bacteria. It has a three-part, segmented, double-stranded RNA genome, totalling ~13.5 kb in length. Φ6 and its relatives have a lipid membrane around their nucleocapsid, a rare trait among bacteriophages. It is a lytic phage, though under certain circumstances has been observed to display a delay in lysis which may be described as a "carrier state".
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.
Icerudivirus is a genus of viruses in the family Rudiviridae. These viruses are non-enveloped, stiff-rod-shaped viruses with linear dsDNA genomes, that infect hyperthermophilic archaea of the species Sulfolobus islandicus. There are three species in the genus.

Lipothrixviridae is a family of viruses in the order Ligamenvirales. Thermophilic archaea in the phylum Thermoproteota serve as natural hosts. There are 11 species in this family, assigned to 4 genera. The genus
Fuselloviridae is a family of viruses. Sulfolobus species, specifically shibatae, solfataricus, and islandicus, serve as natural hosts. There are two genera and nine species in the family. The Fuselloviridae are ubiquitous in high-temperature (≥70 °C), acidic hot springs around the world.

Bottigliavirus is the only genus in the family Ampullaviridae and contains 3 species. Ampullaviridae infect archaea of the genus Acidianus. The name of the family and genus is derived from the Latin word for bottle, ampulla, due to the virions having the shape of a bottle. The family was first described during an investigation of the microbial flora of hot springs in Italy.

Bicaudaviridae is a family of hyperthermophilic archaeal viruses. Members of the genus Acidianus serve as natural hosts. There is only one genus, Bicaudavirus, and one species, Acidianus two-tailed virus, in this family. However, Sulfolobus tengchongensis spindle-shaped viruses 1 and 2 are regarded to belong to this family also.

Haloarcula hispanica pleomorphic virus 1 (HHPV1) is a double stranded DNA virus that infects the halophilic archaeon Haloarcula hispanica. It has a number of unique features unlike any previously described virus.

Halorubrum pleomorphic virus 1 (HRPV-1) is a single stranded DNA virus that infects the species of the archaeal genus Halorubrum. It is unlike any other known virus infecting the archaea with a single stranded DNA genome and an external lipid envelope and is classified in the family Pleolipoviridae, genus Alphapleolipovirus, species Halorubrum virus HRPV1.
Yingchengvirus is a genus of double stranded DNA viruses that infect haloarchaea. The genus was previously named Betasphaerolipovirus.
Alphafusellovirus is a genus of viruses, in the family Fuselloviridae. Species in the genus Sulfolobus serve as natural hosts. There are seven species in this genus.
Tristromaviridae is a family of viruses. Archaea of the genera Thermoproteus and Pyrobaculum serve as natural hosts. Tristromaviridae is the sole family in the order Primavirales. There are two genera and three species in the family.
Spiraviridae is a family of viruses that replicate in hyperthermophilic archaea of the genus Aeropyrum, specifically Aeropyrum pernix. The family contains one genus, Alphaspiravirus, which contains one species, Aeropyrum coil-shaped virus. The virions of Aeropyrum coil-shaped virus (ACV) are non-enveloped and in the shape of hollow cylinders that are formed by a coiling fiber that consists of two intertwining halves of the circular DNA strand inside a capsid. An appendage protrudes from each end of the cylindrical virion. The viral genome is positive-sense, single-stranded DNA ( ssDNA) and encodes for significantly more genes than other known ssDNA viruses. ACV is also unique in that it appears to lack its own enzymes to aid replication, instead likely using the host cell's replisomes. ACV has no known relation to any other archaea-infecting viruses, but it does share its coil-like morphology with some other archaeal viruses, suggesting that such viruses may be an ancient lineage that only infect archaea.
Sulfolobus islandicus rod-shaped virus 2, also referred to as SIRV2, is an archaeal virus whose only known host is the archaeon Sulfolobus islandicus. This virus belongs to the family Rudiviridae. Like other viruses in the family, it is common in geothermal environments.
Pleolipoviridae is a family of DNA viruses that infect archaea.

An archaeal virus is a virus that infects and replicates in archaea, a domain of unicellular, prokaryotic organisms. Archaeal viruses, like their hosts, are found worldwide, including in extreme environments inhospitable to most life such as acidic hot springs, highly saline bodies of water, and at the bottom of the ocean. They have been also found in the human body. The first known archaeal virus was described in 1974 and since then, a large diversity of archaeal viruses have been discovered, many possessing unique characteristics not found in other viruses. Little is known about their biological processes, such as how they replicate, but they are believed to have many independent origins, some of which likely predate the last archaeal common ancestor (LACA).

Thaspiviridae is a family of spindle-shaped viruses that is not assigned to any higher taxonomic ranks. The family contains a single genus, Nitmarvirus, which contains a single species, Nitmarvirus NSV1.

Adnaviria is a realm of viruses that includes archaeal viruses that have a filamentous virion and a linear, double-stranded DNA genome. The genome exists in A-form (A-DNA) and encodes a dimeric major capsid protein (MCP) that contains the SIRV2 fold, a type of alpha-helix bundle containing four helices. The virion consists of the genome encased in capsid proteins to form a helical nucleoprotein complex. For some viruses, this helix is surrounded by a lipid membrane called an envelope. Some contain an additional protein layer between the nucleoprotein helix and the envelope. Complete virions are long and thin and may be flexible or a stiff like a rod.