Related Research Articles
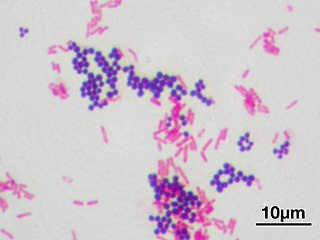
Gram stain or Gram staining, also called Gram's method, is a method of staining used to distinguish and classify bacterial species into two large groups: gram-positive bacteria and gram-negative bacteria. The name comes from the Danish bacteriologist Hans Christian Gram, who developed the technique.

In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall.

Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the gram-staining method of bacterial differentiation. They are characterized by their cell envelopes, which are composed of a thin peptidoglycan cell wall sandwiched between an inner cytoplasmic cell membrane and a bacterial outer membrane.
Peptidoglycan or murein is a polymer consisting of sugars and amino acids that forms a mesh-like layer outside the plasma membrane of most bacteria, forming the cell wall. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is a peptide chain of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. Peptidoglycan is also involved in binary fission during bacterial cell reproduction.

A flagellum is a lash-like appendage that protrudes from the cell body of certain cells termed as flagellates. A flagellate can have one or several flagella. The primary function of a flagellum is that of locomotion, but it also often functions as a sensory organelle, being sensitive to chemicals and temperatures outside the cell.
Mycoplasma pneumoniae is a very small bacterium in the class Mollicutes. It is a human pathogen that causes the disease mycoplasma pneumonia, a form of atypical bacterial pneumonia related to cold agglutinin disease. M. pneumoniae is characterized by the absence of a peptidoglycan cell wall and resulting resistance to many antibacterial agents. The persistence of M. pneumoniae infections even after treatment is associated with its ability to mimic host cell surface composition.

Corynebacterium diphtheriae is the pathogenic bacterium that causes diphtheria. It is also known as the Klebs-Löffler bacillus, because it was discovered in 1884 by German bacteriologists Edwin Klebs (1834–1912) and Friedrich Löffler (1852–1915).
Mollicutes is a class of bacteria distinguished by the absence of a cell wall. The word "Mollicutes" is derived from the Latin mollis, and cutis. Individuals are very small, typically only 0.2–0.3 μm in size and have a very small genome size. They vary in form, although most have sterols that make the cell membrane somewhat more rigid. Many are able to move about through gliding, but members of the genus Spiroplasma are helical and move by twisting. The best-known genus in the Mollicutes is Mycoplasma.
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.
The bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.

Myxococcus xanthus is a gram-negative, rod-shaped species of myxobacteria that exhibits various forms of self-organizing behavior in response to environmental cues. Under normal conditions with abundant food, it exists as a predatory, saprophytic single-species biofilm called a swarm. Under starvation conditions, it undergoes a multicellular development cycle.

Bacteria are a type of biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a number of shapes, ranging from spheres to rods and spirals. Bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of the earth's crust. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised, and only about 27 percent of the bacterial phyla have species that can be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.

A gram-negative bacterial infection is a disease caused by gram-negative bacteria such as E. coli.
Haloplasma contractile is a halophilic, cell wall-less bacterium. It is the only known representative of a deep lineage, and is classified in its own family (Haloplasmataceae) and order (Haloplasmatales), in the class Mollicutes. In terms of genetics, the bacterium Haloplasma contractile contains a dcwgene cluster is responsible for containing all the genes of the organism and promoting peptidoglycan synthesis. Also, MreB/Mbl are specific homologous parts of this bacterium that are vital in the contractility of the cell. In regards to its physical attributes, this organism consists of a spherical body with approximately two protrusions which alternate between straight and contracted forms.

L-form bacteria, also known as L-phase bacteria, L-phase variants, and cell wall-deficient (CWD) bacteria, are strains of bacteria that lack cell walls. They were first isolated in 1935 by Emmy Klieneberger-Nobel, who named them "L-forms" after the Lister Institute in London where she was working.

Gliding motility is a type of translocation used by microorganisms that is independent of propulsive structures such as flagella, pili, and fimbriae. Gliding allows microorganisms to travel along the surface of low aqueous films. The mechanisms of this motility are only partially known.
Chronic Mycoplasma pneumonia and Chlamydia pneumonia infections are associated with the onset and exacerbation of asthma. These microbial infections result in chronic lower airway inflammation, impaired mucociliary clearance, an increase in mucous production and eventually asthma. Furthermore, children who experience severe viral respiratory infections early in life have a high possibility of having asthma later in their childhood. These viral respiratory infections are mostly caused by respiratory syncytial virus (RSV) and human rhinovirus (HRV). Although RSV infections increase the risk of asthma in early childhood, the association between asthma and RSV decreases with increasing age. HRV on the other hand is an important cause of bronchiolitis and is strongly associated with asthma development. In children and adults with established asthma, viral upper respiratory tract infections (URIs), especially HRVs infections, can produce acute exacerbations of asthma. Thus, Chlamydia pneumoniae, Mycoplasma pneumoniae and human rhinoviruses are microbes that play a major role in non-atopic asthma.
Sortases are membrane anchored enzyme that sort these surface proteins onto the bacterial cell surface and anchor them to the peptidoglycan. There are different types of sortases and each catalyse the anchoring of different proteins to cell walls.
Mycoplasma penetrans is a species of Gram-positive bacteria. It is pathogenic, though many infected show no symptoms. It is a sexually transmitted disease, though an infant may be infected during birth.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family
References
- ↑ "Genus Mycoplasma". LPSN. Retrieved 20 February 2018.
- 1 2 3 Adan-Kubo, Jun; Yoshii, Shu-hei; Kono, Hidetoshi; Miyata, Makoto (15 June 2012). "Molecular Structure of Isolated MvspI, a Variable Surface Protein of the Fish Pathogen Mycoplasma mobile". Journal of Bacteriology. 194 (12): 3050–3057. doi:10.1128/JB.00208-12. ISSN 0021-9193. PMC 3370835 . PMID 22447898.
- 1 2 Miyata, Makoto; Hamaguchi, Tasuku (February 2016). "Prospects for the gliding mechanism of Mycoplasma mobile". Current Opinion in Microbiology. 29: 15–21. doi: 10.1016/j.mib.2015.08.010 . PMID 26500189.
- 1 2 3 4 5 Wu, Heng Ning; Miyata, Makoto (24 August 2012). "Whole Surface Image of Mycoplasma mobile, Suggested by Protein Identification and Immunofluorescence Microscopy". Journal of Bacteriology. 194 (21): 5848–5855. doi:10.1128/JB.00976-12. PMC 3486091 . PMID 22923591.
- 1 2 3 4 Morio, Hanako; Kasai, Taishi; Miyata, Makoto; Metcalf, W. W. (15 January 2016). "Gliding Direction of Mycoplasma mobile". Journal of Bacteriology. 198 (2): 283–290. doi:10.1128/JB.00499-15. PMC 4751794 . PMID 26503848.