
The beta sheet is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies.
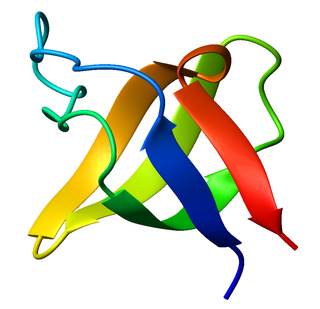
The SRC Homology 3 Domain is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions of proteins involved in the cytoplasmic signaling.

14-3-3 proteins are a family of conserved regulatory molecules that are expressed in all eukaryotic cells. 14-3-3 proteins have the ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors. More than 200 signaling proteins have been reported as 14-3-3 ligands.

Chromosome 1 is the designation for the largest human chromosome. Humans have two copies of chromosome 1, as they do with all of the autosomes, which are the non-sex chromosomes. Chromosome 1 spans about 249 million nucleotide base pairs, which are the basic units of information for DNA. It represents about 8% of the total DNA in human cells.
Chromosome 2 is one of the twenty-three pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 2 is the second-largest human chromosome, spanning more than 242 million base pairs and representing almost eight percent of the total DNA in human cells.
Chromosome 4 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 4 spans more than 193 million base pairs and represents between 6 and 6.5 percent of the total DNA in cells.

In structural biology, a beta-propeller (β-propeller) is a type of all-β protein architecture characterized by 4 to 8 highly symmetrical blade-shaped beta sheets arranged toroidally around a central axis. Together the beta-sheets form a funnel-like active site.

An alpha solenoid is a protein fold composed of repeating alpha helix subunits, commonly helix-turn-helix motifs, arranged in antiparallel fashion to form a superhelix. Alpha solenoids are known for their flexibility and plasticity. Like beta propellers, alpha solenoids are a form of solenoid protein domain commonly found in the proteins comprising the nuclear pore complex. They are also common in membrane coat proteins known as coatomers, such as clathrin, and in regulatory proteins that form extensive protein-protein interactions with their binding partners. Examples of alpha solenoid structures binding RNA and lipids have also been described.

Receptor for activated C kinase 1 (RACK1), also known as guanine nucleotide-binding protein subunit beta-2-like 1 (GNB2L1), is a 35 kDa protein that in humans is encoded by the RACK1 gene.
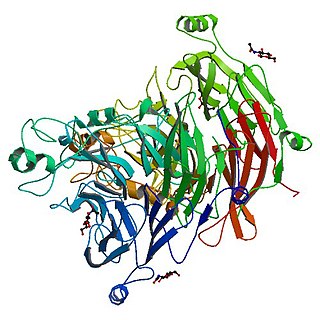
The Sema domain is a structural domain of semaphorins, which are a large family of secreted and transmembrane proteins, some of which function as repellent signals during axon guidance. Sema domains also occur in the hepatocyte growth factor receptor, Plexin-A3 and in viral proteins.

TSBP1 is a protein that in humans is encoded by the TSBP1 gene. TSBP1 was previously known as C6orf10. C6orf10 is an open reading frame on chromosome 6 containing a protein that is ubiquitously expressed at low levels in the adult genome and may play a role during fetal development. C6orf10 has been found to be linked to both neurodegenerative and autoimmune diseases in adults. Expression of this gene is highest in the testis but is also seen in other tissue types such as the brain, lens of the eye and the medulla.

The WD40 repeat is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain.
YWTD repeats are four-stranded beta-propeller repeats found in low-density lipoprotein receptors (LDLR). The six YWTD repeats together fold into a six-bladed beta-propeller. Each blade of the propeller consists of four antiparallel beta-strands; the innermost strand of each blade is labeled 1 and the outermost strand, 4. The sequence repeats are offset with respect to the blades of the propeller, such that any given 40-residue YWTD repeat spans strands 24 of one propeller blade and strand 1 of the subsequent blade. This offset ensures circularization of the propeller because the last strand of the final sequence repeat acts as an innermost strand 1 of the blade that harbors strands 24 from the first sequence repeat. The repeat is found in a variety of proteins that include, vitellogenin receptor from Drosophila melanogaster, low-density lipoprotein (LDL) receptor, preproepidermal growth factor, and nidogen (entactin).
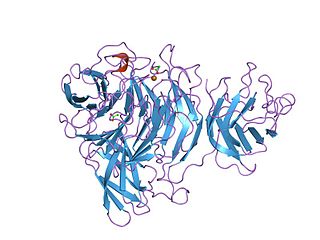
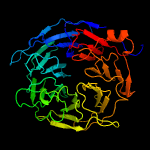
The Kelch motif is a region of protein sequence found widely in proteins from bacteria and eukaryotes. This sequence motif is composed of about 50 amino acid residues which form a structure of a four stranded beta-sheet "blade". This sequence motif is found in between five and eight tandem copies per protein which fold together to form a larger circular solenoid structure called a beta-propeller domain.
The CHASE domain is an extracellular protein domain, which is found in transmembrane receptor from bacteria, lower eukaryotes and plants. It has been named CHASE because of its presence in diverse receptor-like proteins with histidine kinase and nucleotide cyclase domains. The CHASE domain is 200-230 amino acids long and always occurs N-terminally in extracellular or periplasmic locations, followed by an intracellular tail housing diverse enzymatic signalling domains such as histidine kinase, adenyl cyclase, GGDEF-type nucleotide cyclase and EAL-type phosphodiesterase domains, as well as non-enzymatic domains such PAS, GAF, phosphohistidine and response regulatory domains. The CHASE domain is predicted to bind diverse low molecular weight ligands, such as the cytokinin-like adenine derivatives or peptides, and mediate signal transduction through the respective receptors.

In structural and cell biology, the focal adhesion targeting domain is a conserved protein domain that was first identified in focal adhesion kinase (FAK), also known as PTK2 protein tyrosine kinase 2 (PTK2).
In molecular biology, the HAMP domain is an approximately 50-amino acid alpha-helical region that forms a dimeric, four-helical coiled coil. It is found in bacterial sensor and chemotaxis proteins and in eukaryotic histidine kinases. The bacterial proteins are usually integral membrane proteins and part of a two-component signal transduction pathway. One or several copies of the HAMP domain can be found in association with other domains, such as the histidine kinase domain, the bacterial chemotaxis sensory transducer domain, the PAS repeat, the EAL domain, the GGDEF domain, the protein phosphatase 2C-like domain, the guanylate cyclase domain, or the response regulatory domain. In its most common setting, the HAMP domain transmits conformational changes in periplasmic ligand-binding domains to cytoplasmic signalling kinase and methyl-acceptor domains and thus regulates the phosphorylation or methylation activity of homodimeric receptors.

In molecular biology, YopH, N-terminal refers to an evolutionary conserved protein domain. This entry represents the N-terminal domain of YopH protein tyrosine phosphatase (PTP).
Megf8 also known as Multiple Epidermal Growth Factor-like Domains 8, is a protein coding gene that encodes a single pass membrane protein, known to participate in developmental regulation and cellular communication. It is located on chromosome 19 at the 49th open reading frame in humans (19q13.2). There are two isoform constructs known for MEGF8, which differ by a 67 amino acid indel. The isoform 2 splice version is 2785 amino acids long, and predicted to be 296.6 kdal in mass. Isoform 1 is composed of 2845 amino acids and predicted to weigh 303.1 kdal. Using BLAST searches, orthologs were found primarily in mammals, but MEGF8 is also conserved in invertebrates and fishes, and rarely in birds, reptiles, and amphibians. A notably important paralog to multiple epidermal growth factor-like domains 8 is ATRNL1, which is also a single pass transmembrane protein, with several of the same key features and motifs as MEGF8, as indicated by Simple Modular Architecture Research Tool (SMART) which is hosted by the European Molecular Biology Laboratory located in Heidelberg, Germany. MEGF8 has been predicted to be a key player in several developmental processes, such as left-right patterning and limb formation. Currently, researchers have found MEGF8 SNP mutations to be the cause of Carpenter syndrome subtype 2.
NHL Repeat Containing Protein 2, or NHLRC2, is a protein encoded by the NHLRC2 gene.















