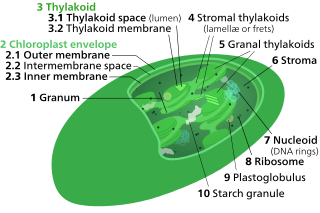
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana. Grana are connected by intergranal or stromal thylakoids, which join granum stacks together as a single functional compartment.

Phycobilisomes are light harvesting antennae of photosystem II in cyanobacteria, red algae and glaucophytes. It was lost in the plastids of green algae / plants (chloroplasts).

Carboxysomes are bacterial microcompartments (BMCs) consisting of polyhedral protein shells filled with the enzymes ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)—the predominant enzyme in carbon fixation and the rate limiting enzyme in the Calvin cycle—and carbonic anhydrase.
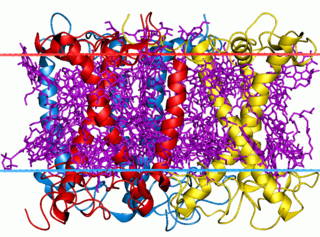
The light-harvesting complex is an array of protein and chlorophyll molecules embedded in the thylakoid membrane of plants and cyanobacteria, which transfer light energy to one chlorophyll a molecule at the reaction center of a photosystem.
A light-harvesting complex consists of a number of chromophores which are complex subunit proteins that may be part of a larger super complex of a photosystem, the functional unit in photosynthesis. It is used by plants and photosynthetic bacteria to collect more of the incoming light than would be captured by the photosynthetic reaction center alone. The light which is captured by the chromophores is capable of exciting molecules from their ground state to a higher energy state, known as the excited state. This excited state does not last very long and is known to be short-lived.
Photoprotection is the biochemical process that helps organisms cope with molecular damage caused by sunlight. Plants and other oxygenic phototrophs have developed a suite of photoprotective mechanisms to prevent photoinhibition and oxidative stress caused by excess or fluctuating light conditions. Humans and other animals have also developed photoprotective mechanisms to avoid UV photodamage to the skin, prevent DNA damage, and minimize the downstream effects of oxidative stress.

Biological pigments, also known simply as pigments or biochromes, are substances produced by living organisms that have a color resulting from selective color absorption. Biological pigments include plant pigments and flower pigments. Many biological structures, such as skin, eyes, feathers, fur and hair contain pigments such as melanin in specialized cells called chromatophores. In some species, pigments accrue over very long periods during an individual's lifespan.
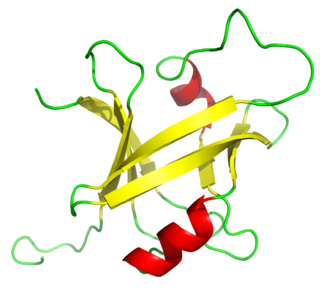
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.
Non-photochemical quenching (NPQ) is a mechanism employed by plants and algae to protect themselves from the adverse effects of high light intensity. It involves the quenching of singlet excited state chlorophylls (Chl) via enhanced internal conversion to the ground state, thus harmlessly dissipating excess excitation energy as heat through molecular vibrations. NPQ occurs in almost all photosynthetic eukaryotes, and helps to regulate and protect photosynthesis in environments where light energy absorption exceeds the capacity for light utilization in photosynthesis.
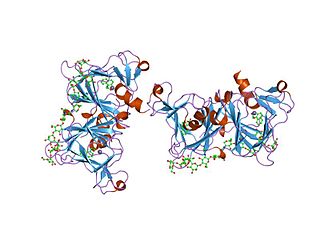
In molecular biology, the auxin binding protein family is a family of proteins which bind auxin. They are located in the lumen of the endoplasmic reticulum (ER). The primary structure of these proteins contains an N-terminal hydrophobic leader sequence of 30-40 amino acids, which could represent a signal for translocation of the protein to the ER. The mature protein comprises around 165 residues, and contains a number of potential N-glycosylation sites. In vitro transport studies have demonstrated co-translational glycosylation. Retention within the lumen of the ER correlates with an additional signal located at the C terminus, represented by the sequence Lys-Asp-Glu-Leu, known to be responsible for preventing secretion of proteins from the lumen of the ER in eukaryotic cells.
Antheraxanthin is a bright yellow accessory pigment found in many organisms that perform photosynthesis. It is a xanthophyll cycle pigment, an oil-soluble alcohol within the xanthophyll subgroup of carotenoids. Antheraxanthin is both a component in and product of the cellular photoprotection mechanisms in photosynthetic green algae, red algae, euglenoids, and plants.
Plastid terminal oxidase or plastoquinol terminal oxidase (PTOX) is an enzyme that resides on the thylakoid membranes of plant and algae chloroplasts and on the membranes of cyanobacteria. The enzyme was hypothesized to exist as a photosynthetic oxidase in 1982 and was verified by sequence similarity to the mitochondrial alternative oxidase (AOX). The two oxidases evolved from a common ancestral protein in prokaryotes, and they are so functionally and structurally similar that a thylakoid-localized AOX can restore the function of a PTOX knockout.

KaiC is a gene belonging to the KaiABC gene cluster that, together, regulate bacterial circadian rhythms, specifically in cyanobacteria. KaiC encodes for the KaiC protein, which interacts with the KaiA and KaiB proteins in a post-translational oscillator (PTO). The PTO is cyanobacteria master clock that is controlled by sequences of phosphorylation of KaiC protein. Regulation of KaiABC expression and KaiABC phosphorylation is essential for cyanobacteria circadian rhythmicity, and is particularly important for regulating cyanobacteria processes such as nitrogen fixation, photosynthesis, and cell division. Studies have shown similarities to Drosophila, Neurospora, and mammalian clock models in that the kaiABC regulation of the cyanobacteria slave circadian clock is also based on a transcription translation feedback loop (TTFL). KaiC protein has both auto-kinase and auto-phosphatase activity and functions as the circadian regulator in both the PTO and the TTFL. KaiC has been found to not only suppress kaiBC when overexpressed, but also suppress circadian expression of all genes in the cyanobacterial genome.
kaiA is a gene in the "kaiABC" gene cluster that plays a crucial role in the regulation of bacterial circadian rhythms, such as in the cyanobacterium Synechococcus elongatus. For these bacteria, regulation of kaiA expression is critical for circadian rhythm, which determines the twenty-four-hour biological rhythm. In addition, KaiA functions with a negative feedback loop in relation with kaiB and KaiC. The kaiA gene makes KaiA protein that enhances phosphorylation of KaiC while KaiB inhibits activity of KaiA.
Iron-starvation-induced protein A, also known as isiA, is a photosynthesis-related chlorophyll-containing protein found in cyanobacteria. It belongs to the chlorophyll-a/b-binding family of proteins, and has been shown to have a photoprotection role in preventing oxidative damage via energy dissipation. It was originally identified under Fe starvation, and thus received the name iron-starvation-induced protein A. However, the protein has more recently been found to respond to a variety of stress conditions such as high irradiance. It can aggregate with carotenoids and form rings around the PSI reaction center complexes to aid in photoprotective energy dissipation.
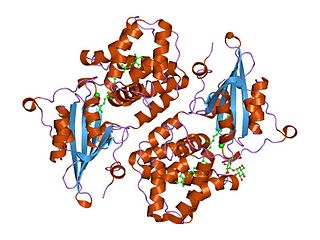
Orange carotenoid protein (OCP) is a water-soluble protein which plays a role in photoprotection in diverse cyanobacteria. It is the only photoactive protein known to use a carotenoid as the photoresponsive chromophore. The protein consists of two domains, with a single keto-carotenoid molecule non-covalently bound between the two domains. It is a very efficient quencher of excitation energy absorbed by the primary light-harvesting antenna complexes of cyanobacteria, the phycobilisomes. The quenching is induced by blue-green light. It is also capable of preventing oxidative damage by directly scavenging singlet oxygen (1O2).
In photosynthesis, state transitions are rearrangements of the photosynthetic apparatus which occur on short time-scales. The effect is prominent in cyanobacteria, whereby the phycobilisome light-harvesting antenna complexes alter their preference for transfer of excitation energy between the two reaction centers, PS I and PS II. This shift helps to minimize photodamage caused by reactive oxygen species (ROS) under stressful conditions such as high light, but may also be used to offset imbalances between the rates of generating reductant and ATP.
3′-Hydroxyechinenone is a keto-carotenoid pigment found in cyanobacteria and microalgae. Carotenoids belong to a larger class of phytochemicals known as terpenoids. The chemical formula of canthaxanthin is C40H54O2. It is found non-covalently bound in the orange carotenoid protein (OCP), which is a soluble protein involved in photoprotection and non-photochemical quenching of photosynthesis.
Fluorescence recovery protein (FRP) is a small protein involved in regulating non-photochemical quenching in cyanobacteria. It prevents accumulation of the red photoactivated form of orange carotenoid protein (OCP), thereby reducing the amount of fluorescence quenching that occurs between the OCP and the phycobilisome antenna complexes. It interacts with the C-terminal domain of OCP, which shares homology with the NTF2 superfamily.
David W. Krogmann was an American biologist and a professor of biochemistry at Purdue University. He is known for his work in photosynthesis in chloroplasts and cyanobacteria.








