
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.

5α-Reductases, also known as 3-oxo-5α-steroid 4-dehydrogenases, are enzymes involved in steroid metabolism. They participate in three metabolic pathways: bile acid biosynthesis, androgen and estrogen metabolism. There are three isozymes of 5α-reductase encoded by the genes SRD5A1, SRD5A2, and SRD5A3.
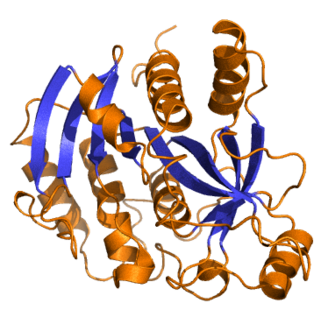
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction

Queuine (Q) is a hypermodified nucleobase found in the first position of the anticodon of tRNAs specific for Asn, Asp, His, and Tyr, in most eukaryotes and prokaryotes. Because it is utilized by all eukaryotes but produced exclusively by bacteria, it is a putative vitamin.
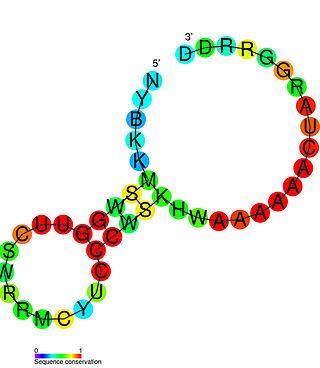
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.
In enzymology, a dihydrokaempferol 4-reductase (EC 1.1.1.219) is an enzyme that catalyzes the chemical reaction

Sepiapterin reductase is an enzyme that in humans is encoded by the SPR gene.
A glutamyl-tRNA reductase (EC 1.2.1.70) is an enzyme that catalyzes the chemical reaction

[Methionine synthase] reductase, or Methionine synthase reductase, encoded by the gene MTRR, is an enzyme that is responsible for the reduction of methionine synthase inside human body. This enzyme is crucial for maintaining the one carbon metabolism, specifically the folate cycle. The enzyme employs one coenzyme, flavoprotein.
In enzymology, a queuine tRNA-ribosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a tRNA-queuosine beta-mannosyltransferase is an enzyme that catalyzes the chemical reaction

C9orf64 is a gene located on chromosome 9, that in humans encodes the protein queuosine salvage protein. The function and biological process of the queuosine salvage protein is a queuosine-nucleotide N-glycosylase/hydrolase (QNG1) that releases queuine from Q-5'-monophosphate, and this activity is required for the salvage of queuine from exogenous Queuosine by S. pombe and HeLa cells. Some evidence from orthologs indicates it may be involved in tRNA processing and recycling. The most common mRNA contains 4 coding exons, and it has 2 additional alternatively spliced exons. C9orf64 has been found in 5 different splice variants.
Geranylgeranyl diphosphate reductase (EC 1.3.1.83, geranylgeranyl reductase, CHL P) is an enzyme with systematic name geranylgeranyl-diphosphate:NADP+ oxidoreductase. This enzyme catalises the following chemical reaction
FMN reductase (NADPH) (EC 1.5.1.38, FRP, flavin reductase P, SsuE) is an enzyme with systematic name FMNH2:NADP+ oxidoreductase. This enzyme catalyses the following chemical reaction:
TRNA-guanine15 transglycosylase is an enzyme with systematic name tRNA-guanine15:7-cyano-7-carbaguanine tRNA-D-ribosyltransferase. This enzyme catalyses the following chemical reaction
S-Adenosylmethionine:tRNA ribosyltransferase-isomerase is an enzyme with systematic name S-adenosyl-L-methionine:7-aminomethyl-7-deazaguanosine ribosyltransferase . This enzyme catalyses the following chemical reaction
Archaeosine synthase is an enzyme with systematic name L-glutamine:7-cyano-7-carbaguanine aminotransferase. This enzyme catalyses the following chemical reaction
7-cyano-7-deazaguanine synthase (EC 6.3.4.20, preQ0 synthase, 7-cyano-7-carbaguanine synthase, queC (gene)) is an enzyme with systematic name 7-carboxy-7-carbaguanine:ammonia ligase (ADP-forming). This enzyme catalyses the following chemical reaction









