
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

Omenn syndrome is an autosomal recessive severe combined immunodeficiency. It is associated with hypomorphic missense mutations in immunologically relevant genes of T-cells such as recombination activating genes, Interleukin-7 receptor-α (IL7Rα), DCLRE1C-Artemis, RMRP-CHH, DNA-Ligase IV, common gamma chain, WHN-FOXN1, ZAP-70 and complete DiGeorge syndrome. It is fatal without treatment.
V(D)J recombination is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire of antibodies/immunoglobulins and T cell receptors (TCRs) found in B cells and T cells, respectively. The process is a defining feature of the adaptive immune system.
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or be triggered by a specific external stimulus. It is implemented both in eukaryotic and prokaryotic systems. The Cre-lox recombination system has been particularly useful to help neuroscientists to study the brain in which complex cell types and neural circuits come together to generate cognition and behaviors. NIH Blueprint for Neuroscience Research has created several hundreds of Cre driver mouse lines which are currently used by the worldwide neuroscience community.

In genetics, Flp-FRT recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions in vivo. It is analogous to Cre-lox recombination but involves the recombination of sequences between short flippase recognition target (FRT) sites by the recombinase flippase (Flp) derived from the 2 µ plasmid of baker's yeast Saccharomyces cerevisiae.
Allelic exclusion is a process by which only one allele of a gene is expressed while the other allele is silenced. This phenomenon is most notable for playing a role in the development of B lymphocytes, where allelic exclusion allows for each mature B lymphocyte to express only one type of immunoglobulin. This subsequently results in each B lymphocyte being able to recognize only one antigen. This is significant as the co-expression of both alleles in B lymphocytes is associated with autoimmunity and the production of autoantibodies.
The recombination-activating genes (RAGs) encode parts of a protein complex that plays important roles in the rearrangement and recombination of the genes encoding immunoglobulin and T cell receptor molecules. There are two recombination-activating genes RAG1 and RAG2, whose cellular expression is restricted to lymphocytes during their developmental stages. The enzymes encoded by these genes, RAG-1 and RAG-2, are essential to the generation of mature B cells and T cells, two types of lymphocyte that are crucial components of the adaptive immune system.

Recombination activating gene 1 also known as RAG-1 is a protein that in humans is encoded by the RAG1 gene.
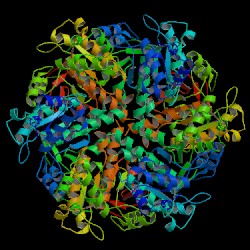
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene RAD51. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to the bacterial RecA, Archaeal RadA and yeast Rad51. The protein is highly conserved in most eukaryotes, from yeast to humans.

Serine/threonine-protein kinase ATR also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1) is an enzyme that, in humans, is encoded by the ATR gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity.

Nibrin, also known as NBN or NBS1, is a protein which in humans is encoded by the NBN gene.
Conditional gene knockout is a technique used to eliminate a specific gene in a certain tissue, such as the liver. This technique is useful to study the role of individual genes in living organisms. It differs from traditional gene knockout because it targets specific genes at specific times rather than being deleted from beginning of life. Using the conditional gene knockout technique eliminates many of the side effects from traditional gene knockout. In traditional gene knockout, embryonic death from a gene mutation can occur, and this prevents scientists from studying the gene in adults. Some tissues cannot be studied properly in isolation, so the gene must be inactive in a certain tissue while remaining active in others. With this technology, scientists are able to knockout genes at a specific stage in development and study how the knockout of a gene in one tissue affects the same gene in other tissues.

Ku70 is a protein that, in humans, is encoded by the XRCC6 gene.

Artemis is a protein that in humans is encoded by the DCLRE1C gene.

Nuclear factor of activated T-cells 5, also known as NFAT5 and sometimes TonEBP, is a human gene that encodes a transcription factor that regulates the expression of genes involved in the osmotic stress.

High-mobility group protein B2 also known as high-mobility group protein 2 (HMG-2) is a protein that in humans is encoded by the HMGB2 gene.

Meiotic recombination protein DMC1/LIM15 homolog is a protein that in humans is encoded by the DMC1 gene.

Crossover junction endonuclease EME1 is an enzyme that in humans is encoded by the EME1 gene. It forms a complex with MUS81 which resolves Holliday junctions. In mammalian cells the EME1/MUS81 protein complex is redundant for DNA damage repair with GEN1 endonuclease. In mice, EME1/MUS81 and GEN1 redundantly contribute to Holliday junction processing. When homozygous mutations of Gen1 and Eme1 were combined in mice the result was synthetic lethality at an early embryonic stage. Homozygosity for Gen1 mutations did not cause a DNA repair deficiency in mice. But when mice were both homozygous mutant for Gen1 and also heterozyous for an Emc1 mutation, they showed increased sensitivity to DNA damaging agents. This finding, indicated a redundant role of GEN1 and EME1 in DNA repair. Gen1 and Emc1 were also shown to have redundant roles in meiotic recombination.
Recombination signal sequences are conserved sequences of noncoding DNA that are recognized by the RAG1/RAG2 enzyme complex during V(D)J recombination in immature B cells and T cells. Recombination signal sequences guide the enzyme complex to the V, D, and J gene segments that will undergo recombination during the formation of the heavy and light-chain variable regions in T-cell receptors and immunoglobulin molecules.

In genetics, floxing refers to the sandwiching of a DNA sequence between two lox P sites. The terms are constructed upon the phrase "flanking/flanked by LoxP". Recombination between LoxP sites is catalysed by Cre recombinase. Floxing a gene allows it to be deleted, translocated or inverted in a process called Cre-Lox recombination. The floxing of genes is essential in the development of scientific model systems as it allows researchers to have spatial and temporal alteration of gene expression. Moreover, animals such as mice can be used as models to study human disease. Therefore, Cre-lox system can be used in mice to manipulate gene expression in order to study human diseases and drug development. For example, using the Cre-lox system, researchers can study oncogenes and tumor suppressor genes and their role in development and progression of cancer in mice models.

















