Related Research Articles

Superoxide dismutase (SOD, EC 1.15.1.1) is an enzyme that alternately catalyzes the dismutation (or partitioning) of the superoxide (O−
2) radical into ordinary molecular oxygen (O2) and hydrogen peroxide (H
2O
2). Superoxide is produced as a by-product of oxygen metabolism and, if not regulated, causes many types of cell damage. Hydrogen peroxide is also damaging and is degraded by other enzymes such as catalase. Thus, SOD is an important antioxidant defense in nearly all living cells exposed to oxygen. One exception is Lactobacillus plantarum and related lactobacilli, which use a different mechanism to prevent damage from reactive O−
2.

Xanthophylls are yellow pigments that occur widely in nature and form one of two major divisions of the carotenoid group; the other division is formed by the carotenes. The name is from Greek xanthos and phyllon, due to their formation of the yellow band seen in early chromatography of leaf pigments.

Reactive oxygen species (ROS) are highly reactive chemicals formed from O2. Examples of ROS include peroxides, superoxide, hydroxyl radical, singlet oxygen, and alpha-oxygen.

Obligate anaerobes are microorganisms killed by normal atmospheric concentrations of oxygen (20.95% O2). Oxygen tolerance varies between species, with some species capable of surviving in up to 8% oxygen, while others lose viability in environments with an oxygen concentration greater than 0.5%.

Photosystems are functional and structural units of protein complexes involved in photosynthesis. Together they carry out the primary photochemistry of photosynthesis: the absorption of light and the transfer of energy and electrons. Photosystems are found in the thylakoid membranes of plants, algae, and cyanobacteria. These membranes are located inside the chloroplasts of plants and algae, and in the cytoplasmic membrane of photosynthetic bacteria. There are two kinds of photosystems: PSI and PSII.
Respiratory burst is the rapid release of the reactive oxygen species (ROS), superoxide anion and hydrogen peroxide, from different cell types.

Phycocyanin is a pigment-protein complex from the light-harvesting phycobiliprotein family, along with allophycocyanin and phycoerythrin. It is an accessory pigment to chlorophyll. All phycobiliproteins are water-soluble, so they cannot exist within the membrane like carotenoids can. Instead, phycobiliproteins aggregate to form clusters that adhere to the membrane called phycobilisomes. Phycocyanin is a characteristic light blue color, absorbing orange and red light, particularly near 620 nm, and emits fluorescence at about 650 nm. Allophycocyanin absorbs and emits at longer wavelengths than phycocyanin C or phycocyanin R. Phycocyanins are found in cyanobacteria. Phycobiliproteins have fluorescent properties that are used in immunoassay kits. Phycocyanin is from the Greek phyco meaning “algae” and cyanin is from the English word “cyan", which conventionally means a shade of blue-green and is derived from the Greek “kyanos" which means a somewhat different color: "dark blue". The product phycocyanin, produced by Aphanizomenon flos-aquae and Spirulina, is for example used in the food and beverage industry as the natural coloring agent 'Lina Blue' or 'EXBERRY Shade Blue' and is found in sweets and ice cream. In addition, fluorescence detection of phycocyanin pigments in water samples is a useful method to monitor cyanobacteria biomass.

Carotenoid oxygenases are a family of enzymes involved in the cleavage of carotenoids to produce, for example, retinol, commonly known as vitamin A. This family includes an enzyme known as RPE65 which is abundantly expressed in the retinal pigment epithelium where it catalyzed the formation of 11-cis-retinol from all-trans-retinyl esters.

Biological pigments, also known simply as pigments or biochromes, are substances produced by living organisms that have a color resulting from selective color absorption. Biological pigments include plant pigments and flower pigments. Many biological structures, such as skin, eyes, feathers, fur and hair contain pigments such as melanin in specialized cells called chromatophores. In some species, pigments accrue over very long periods during an individual's lifespan.

Superoxide dismutase [Cu-Zn] also known as superoxide dismutase 1 or hSod1 is an enzyme that in humans is encoded by the SOD1 gene, located on chromosome 21. SOD1 is one of three human superoxide dismutases. It is implicated in apoptosis and familial amyotrophic lateral sclerosis.
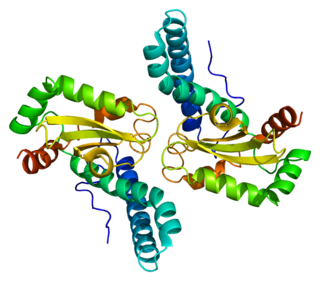
Superoxide dismutase 2, mitochondrial (SOD2), also known as manganese-dependent superoxide dismutase (MnSOD), is an enzyme which in humans is encoded by the SOD2 gene on chromosome 6. A related pseudogene has been identified on chromosome 1. Alternative splicing of this gene results in multiple transcript variants. This gene is a member of the iron/manganese superoxide dismutase family. It encodes a mitochondrial protein that forms a homotetramer and binds one manganese ion per subunit. This protein binds to the superoxide byproducts of oxidative phosphorylation and converts them to hydrogen peroxide and diatomic oxygen. Mutations in this gene have been associated with idiopathic cardiomyopathy (IDC), premature aging, sporadic motor neuron disease, and cancer.

Pyocyanin (PCN−) is one of the many toxins produced and secreted by the Gram negative bacterium Pseudomonas aeruginosa. Pyocyanin is a blue, secondary metabolite with the ability to oxidise and reduce other molecules and therefore kill microbes competing against P. aeruginosa as well as mammalian cells of the lungs which P. aeruginosa has infected during cystic fibrosis. Since pyocyanin is a zwitterion at blood pH, it is easily able to cross the cell membrane. There are three different states in which pyocyanin can exist: oxidized, monovalently reduced or divalently reduced. Mitochondria play an important role in the cycling of pyocyanin between its redox states. Due to its redox-active properties, pyocyanin generates reactive oxygen species.
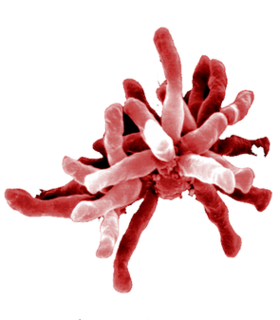
Rhodopseudomonas palustris is a rod-shaped, Gram-negative purple nonsulfur bacterium, notable for its ability to switch between four different modes of metabolism.
All living cells produce reactive oxygen species (ROS) as a byproduct of metabolism. ROS are reduced oxygen intermediates that include the superoxide radical (O2−) and the hydroxyl radical (OH•), as well as the non-radical species hydrogen peroxide (H2O2). These ROS are important in the normal functioning of cells, playing a role in signal transduction and the expression of transcription factors. However, when present in excess, ROS can cause damage to proteins, lipids and DNA by reacting with these biomolecules to modify or destroy their intended function. As an example, the occurrence of ROS have been linked to the aging process in humans, as well as several other diseases including Alzheimer's, rheumatoid arthritis, Parkinson's, and some cancers. Their potential for damage also makes reactive oxygen species useful in direct protection from invading pathogens, as a defense response to physical injury, and as a mechanism for stopping the spread of bacteria and viruses by inducing programmed cell death.
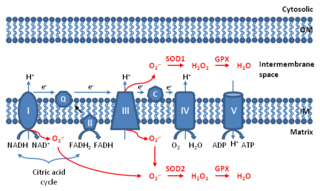
Mitochondrial ROS are reactive oxygen species (ROS) that are produced by mitochondria. Generation of mitochondrial ROS mainly takes place at the electron transport chain located on the inner mitochondrial membrane during the process of oxidative phosphorylation. Leakage of electrons at complex I and complex III from electron transport chains leads to partial reduction of oxygen to form superoxide. Subsequently, superoxide is quickly dismutated to hydrogen peroxide by two dismutases including superoxide dismutase 2 (SOD2) in mitochondrial matrix and superoxide dismutase 1 (SOD1) in mitochondrial intermembrane space. Collectively, both superoxide and hydrogen peroxide generated in this process are considered as mitochondrial ROS.
Oxidation response is stimulated by a disturbance in the balance between the production of reactive oxygen species and antioxidant responses, known as oxidative stress. Active species of oxygen naturally occur in aerobic cells and have both intracellular and extracellular sources. These species, if not controlled, damage all components of the cell, including proteins, lipids and DNA. Hence cells need to maintain a strong defense against the damage. The following table gives an idea of the antioxidant defense system in bacterial system.
Halorhodospira halophila is a species of Halorhodospira distinguished by its ability to grow optimally in an environment of 15–20% salinity. It was formerly called Ectothiorhodospira halophila. It is an anaerobic, rod-shaped Gram-negative bacterium. H. halophila has a flagellum.

Azotobacter chroococcum is a bacterium that has the ability to fix atmospheric nitrogen. It was discovered by Martinus Beijerinck in 1901, and was the first aerobic, free-living nitrogen fixer discovered. A. chroococcum could be useful for nitrogen fixation in crops as a biofertilizer, fungicide, and nutrient indicator, and in bioremediation.
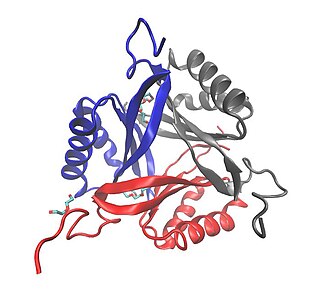
SbtB, which stands for sodium-bicarbonate-transporter B, is a protein found in bacteria. This small soluble protein has been classified as a new member of the P-II family that is involved in signal transduction. This protein has been demonstrated to participate in numerous processes including carbon sensing mechanisms in cyanobacteria.
Thioflavicoccus is a Gram-negative, obligately phototrophic, strictly anaerobic and motile genus of bacteria from the family of Chromatiaceae with one known species.
References
- ↑ LPSN lpsn.dsmz.de
- ↑ Gille de Luca; Mohamed Barakat; Philippe Ortet; Sylvain Fochesato; Cécile Jourlin-Castelli; Mireille Ansaldi; Béatrice Py; Gwennaele Fichant; Pedro M. Coutinho; Romé Voulhoux; Olivier Bastien; Eric Maréchal; Bernard Henrissat; Yves Quentin; Philippe Noirot; Alain Filloux; Vincent Méjean; Michael S. DuBow; Frédéric Barras; Valérie Barbe; Jean Weissenbach; Irina Mihalcescu; André Verméglio; Wafa Achouak; Thierry Heulin (September 2011). "The cyst-dividing bacterium Ramlibacter tataouinensis TTB310 genome reveals a well-stocked toolbox for adaptation to a desert environment". PLoS ONE . 6 (9): e23784. Bibcode:2011PLoSO...623784D. doi: 10.1371/journal.pone.0023784 . PMC 3164672 . PMID 21912644.