RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.
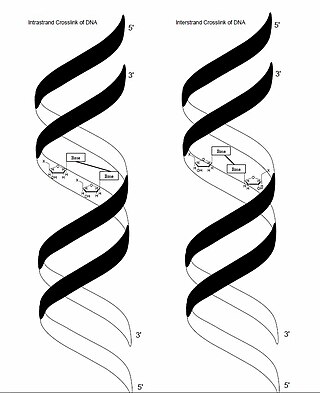
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.

DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. The gene is commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH1 have also been studied in other organisms including mouse and the budding yeast Saccharomyces cerevisiae.

Replication protein A 70 kDa DNA-binding subunit is a protein that in humans is encoded by the RPA1 gene.

Mismatch repair endonuclease PMS2 is an enzyme that in humans is encoded by the PMS2 gene.

DNA excision repair protein ERCC-1 is a protein that in humans is encoded by the ERCC1 gene. Together with ERCC4, ERCC1 forms the ERCC1-XPF enzyme complex that participates in DNA repair and DNA recombination.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.

Exonuclease 1 is an enzyme that in humans is encoded by the EXO1 gene.
ERCC4 is a protein designated as DNA repair endonuclease XPF that in humans is encoded by the ERCC4 gene. Together with ERCC1, ERCC4 forms the ERCC1-XPF enzyme complex that participates in DNA repair and DNA recombination.

DNA repair and recombination protein RAD54-like is a protein that in humans is encoded by the RAD54L gene.

Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the MUS81 gene.

DNA mismatch repair protein Mlh3 is a protein that in humans is encoded by the MLH3 gene.

DNA cross-link repair 1A protein is a protein that in humans is encoded by the DCLRE1A gene.

DNA cross-link repair 1B protein is a protein that in humans is encoded by the DCLRE1B gene.

Fanconi anemia, complementation group M, also known as FANCM is a human gene. It is an emerging target in cancer therapy, in particular cancers with specific genetic deficiencies.

SLX4 is a protein involved in DNA repair, where it has important roles in the final steps of homologous recombination. Mutations in the gene are associated with the disease Fanconi anemia.
Telomere-binding proteins function to bind telomeric DNA in various species. In particular, telomere-binding protein refers to TTAGGG repeat binding factor-1 (TERF1) and TTAGGG repeat binding factor-2 (TERF2). Telomere sequences in humans are composed of TTAGGG sequences which provide protection and replication of chromosome ends to prevent degradation. Telomere-binding proteins can generate a T-loop to protect chromosome ends. TRFs are double-stranded proteins which are known to induce bending, looping, and pairing of DNA which aids in the formation of T-loops. They directly bind to TTAGGG repeat sequence in the DNA. There are also subtelomeric regions present for regulation. However, in humans, there are six subunits forming a complex known as shelterin.

GEN1, Holliday junction 5' flap endonuclease is a protein that in humans is encoded by the GEN1 gene.


















