
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.
Topoisomerases are enzymes that participate in the overwinding or underwinding of DNA. The winding problem of DNA arises due to the intertwined nature of its double-helical structure. During DNA replication and transcription, DNA becomes overwound ahead of a replication fork. If left unabated, this torsion would eventually stop the ability of DNA or RNA polymerases involved in these processes to continue down the DNA strand.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions needing it to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

The signal recognition particle RNA, is part of the signal recognition particle (SRP) ribonucleoprotein complex. SRP recognizes the signal peptide and binds to the ribosome, halting protein synthesis. SRP-receptor is a protein that is embedded in a membrane, and which contains a transmembrane pore. When the SRP-ribosome complex binds to SRP-receptor, SRP releases the ribosome and drifts away. The ribosome resumes protein synthesis, but now the protein is moving through the SRP-receptor transmembrane pore.

Tetraloops are a type of four-base hairpin loop motifs in RNA secondary structure that cap many double helices. There are many variants of the tetraloop. The published ones include ANYA, CUYG, GNRA, UNAC and UNCG.

60S ribosomal protein L41 is a protein that in humans is encoded by the RPL41 gene.

AIDS is caused by the human immunodeficiency virus (HIV). Individuals with HIV have what is referred to as a "HIV infection". When infected semen, vaginal secretions, or blood come in contact with the mucous membranes or broken skin of an uninfected person, HIV may be transferred to the uninfected person, causing another infection. Additionally, HIV can also be passed from infected pregnant women to their uninfected baby during pregnancy and/or delivery, or via breastfeeding. As a result of HIV infection, a portion of these individuals will progress and go on to develop clinically significant AIDS.

A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.

Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional tertiary structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structure motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized.

The c4 antisense RNA is a non-coding RNA used by certain phages that infect bacteria. It was initially identified in the P1 and P7 phages of E. coli. The identification of c4 antisense RNAs solved the mystery of the mechanism for regulation of the ant gene, which is an anti-repressor.

The Cyano-2 RNA motif is a conserved RNA structure identified by bioinformatics. Cyano-2 RNAs are found in Cyanobacterial species classified within the genus Synechococcus. Many terminal loops in the two conserved stem-loops contain the nucleotide sequence GCGA, and these sequences might in some cases form stable GNRA tetraloops. Since the two stem-loops are somewhat distant from one another it is possible that they represent two independent non-coding RNAs that are often or always co-transcribed. The region one thousand base pairs upstream of predicted Cyano-2 RNAs is usually devoid of annotated features such as RNA or protein-coding genes. This absence of annotated genes within one thousand base pairs is relatively unusual within bacteria.

The potC RNA motif is a conserved RNA structure discovered using bioinformatics. The RNA is detected only in genome sequences derived from DNA that was extracted from uncultivated marine bacteria. Thus, this RNA is present in environmental samples, but not yet found in any cultivated organism. potC RNAs are located in the presumed 5' untranslated regions of genes predicted to encode either membrane transport proteins or peroxiredoxins. Therefore, it was hypothesized that potC RNAs are cis-regulatory elements, but their detailed function is unknown.

The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.

The radC RNA motif is a conserved RNA structure identified by bioinformatics. The radC RNA motif is found in certain bacteria where it is consistent located in the presumed 5' untranslated regions of genes whose encoded proteins bind DNA are interact with other proteins that bind DNA. These proteins include integrases, methyltransferases that might methylate DNA, proteins that inhibit restriction enzymes and radC genes. Although radC genes were thought to encode DNA repair proteins, this conclusion was based on mutation data that was later shown to affect a different gene. However, it is still possible that radC genes play some DNA-related role. No radC RNAs have been detected in any purified phage whose sequence was available as of 2010, although integrases are often used by phages.

Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary.

The Whalefall-1 RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. Structurally, the motif consists of two stem-loops, the second of which is often terminated by a CUUG tetraloop, which is an energetically favorable RNA sequence. Whalefall-1 RNAs are found only in DNA extracted from uncultivated bacteria found on whale fall, i.e., a whale carcass. As of 2010, Whalefall-1 RNAs have not been detected in any known, cultivated species of bacteria, and are thus one of several RNAs present in environmental samples.
The first human immunodeficiency virus (HIV) case was reported in the United States in the early 1980s. Many drugs have been discovered to treat the disease but mutations in the virus and resistance to the drugs make development difficult. Integrase is a viral enzyme that integrates retroviral DNA into the host cell genome. Integrase inhibitors are a new class of drugs used in the treatment of HIV. The first integrase inhibitor, raltegravir, was approved in 2007 and other drugs were in clinical trials in 2011.
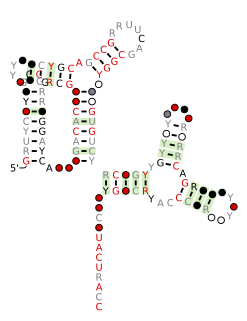
The xerDC RNA motif is a conserved RNA structure that was discovered by bioinformatics. xerDC motif RNAs are found in Clostridia.
















