
Intermediate filaments (IFs) are cytoskeletal structural components found in the cells of vertebrates, and many invertebrates. Homologues of the IF protein have been noted in an invertebrate, the cephalochordate Branchiostoma.
A coiled coil is a structural motif in proteins in which 2–7 alpha-helices are coiled together like the strands of a rope. They have been found in roughly 5-10% of proteins and have a variety of functions. They are one of the most widespread motifs found in protein-protein interactions. To aid protein study, several tools have been developed to predict coiled-coils in protein structures. Many coiled coil-type proteins are involved in important biological functions, such as the regulation of gene expression — e.g., transcription factors. Notable examples are the oncoproteins c-Fos and c-Jun, as well as the muscle protein tropomyosin.

Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a residue, which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, cryo-electron microscopy (cryo-EM) and dual polarisation interferometry, to determine the structure of proteins.

In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins.

Pleckstrin homology domain or (PHIP) is a protein domain of approximately 120 amino acids that occurs in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton.

Desmoplakin is a protein in humans that is encoded by the DSP gene. Desmoplakin is a critical component of desmosome structures in cardiac muscle and epidermal cells, which function to maintain the structural integrity at adjacent cell contacts. In cardiac muscle, desmoplakin is localized to intercalated discs which mechanically couple cardiac cells to function in a coordinated syncytial structure. Mutations in desmoplakin have been shown to play a role in dilated cardiomyopathy and arrhythmogenic right ventricular cardiomyopathy, where it may present with acute myocardial injury; striate palmoplantar keratoderma, Carvajal syndrome and paraneoplastic pemphigus.
Actinin is a microfilament protein. The functional protein is an anti-parallel dimer, which cross-links the thin filaments in adjacent sarcomeres, and therefore coordinates contractions between sarcomeres in the horizontal axis. Alpha-actinin is a part of the spectrin superfamily. This superfamily is made of spectrin, dystrophin, and their homologous and isoforms. In non-muscle cells, it is found by the actin filaments and at the adhesion sites.The lattice like arrangement provides stability to the muscle contractile apparatus. Specifically, it helps bind actin filaments to the cell membrane. There is a binding site at each end of the rod and with bundles of actin filaments.

In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.
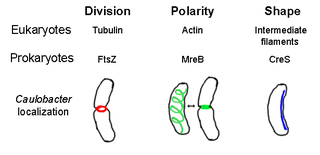
The prokaryotic cytoskeleton is the collective name for all structural filaments in prokaryotes. It was once thought that prokaryotic cells did not possess cytoskeletons, but advances in visualization technology and structure determination led to the discovery of filaments in these cells in the early 1990s. Not only have analogues for all major cytoskeletal proteins in eukaryotes been found in prokaryotes, cytoskeletal proteins with no known eukaryotic homologues have also been discovered. Cytoskeletal elements play essential roles in cell division, protection, shape determination, and polarity determination in various prokaryotes.

Dynactin is a 23 subunit protein complex that acts as a co-factor for the microtubule motor cytoplasmic dynein-1. It is built around a short filament of actin related protein-1 (Arp1).

Alpha II-spectrin, also known as Spectrin alpha chain, brain is a protein that in humans is encoded by the SPTAN1 gene. Alpha II-spectrin is expressed in a variety of tissues, and is highly expressed in cardiac muscle at Z-disc structures, costameres and at the sarcolemma membrane. Mutations in alpha II-spectrin have been associated with early infantile epileptic encephalopathy-5, and alpha II-spectrin may be a valuable biomarker for Guillain–Barré syndrome and infantile congenital heart disease.

Spectrin beta chain, brain 1 is a protein that in humans is encoded by the SPTBN1 gene.

Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 is a protein that in humans is encoded by the MACF1 gene.
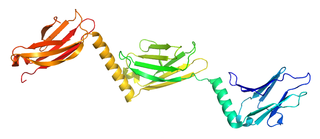
Myomesin-1 is a protein that in humans is encoded by the MYOM1 gene. Myomesin-1 is expressed in muscle cells and functions to stabilize the three-dimensional conformation of the thick filament. Embryonic forms of Myomesin-1 have been detected in dilated cardiomyopathy.

In molecular biology, proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of conformations. Transitions between these states occur on a variety of length scales and time scales , and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis.

The WW domain is a modular protein domain that mediates specific interactions with protein ligands. This domain is found in a number of unrelated signaling and structural proteins and may be repeated up to four times in some proteins. Apart from binding preferentially to proteins that are proline-rich, with particular proline-motifs, [AP]-P-P-[AP]-Y, some WW domains bind to phosphoserine- and phosphothreonine-containing motifs.
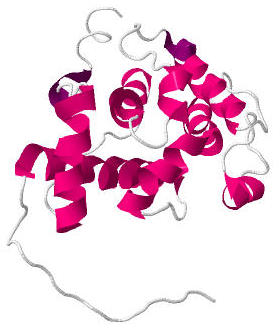
Calponin homology domain (or CH domain) is a family of actin binding domains found in both cytoskeletal proteins and signal transduction proteins. The domain is about 100 amino acids in length and is composed of four alpha helices. It comprises the following groups of actin-binding domains:

In molecular biology, protein fold classes are broad categories of protein tertiary structure topology. They describe groups of proteins that share similar amino acid and secondary structure proportions. Each class contains multiple, independent protein superfamilies.

Hartmut Oschkinat is a German structural biologist and professor for chemistry at the Free University of Berlin. His research focuses on the study of biological systems with solid-state nuclear magnetic resonance.

An array of protein tandem repeats is defined as several adjacent copies having the same or similar sequence motifs. These periodic sequences are generated by internal duplications in both coding and non-coding genomic sequences. Repetitive units of protein tandem repeats are considerably diverse, ranging from the repetition of a single amino acid to domains of 100 or more residues.


















