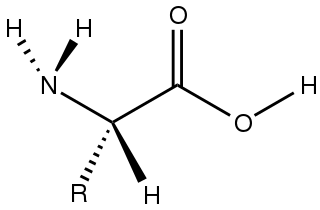
Amino acids are organic compounds that contain amino and carboxylate −CO−2 functional groups, along with a side chain specific to each amino acid. The elements present in every amino acid are carbon (C), hydrogen (H), oxygen (O), and nitrogen (N); in addition sulfur (S) is present in the side chains of cysteine and methionine, and selenium (Se) in the less common amino acid selenocysteine. More than 500 naturally occurring amino acids are known to constitute monomer units of peptides, including proteins, as of 2020

Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.
Peptides are short chains of amino acids linked by peptide bonds. Chains of fewer than ten or fifteen amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides.

Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences.
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group -NH
2 but is rather a secondary amine. The secondary amine nitrogen is in the protonated NH2+ form under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG).
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The epitope is the specific piece of the antigen to which an antibody binds. The part of an antibody that binds to the epitope is called a paratope. Although epitopes are usually non-self proteins, sequences derived from the host that can be recognized are also epitopes.

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length, that serves as the physical link between the mRNA and the amino acid sequence of proteins. Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a nonsense codon in the transcribed mRNA, and in a truncated, incomplete, and usually nonfunctional protein product. The functional effect of a nonsense mutation depends on the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected.
Protein sequencing is the practical process of determining the amino acid sequence of all or part of a protein or peptide. This may serve to identify the protein or characterize its post-translational modifications. Typically, partial sequencing of a protein provides sufficient information to identify it with reference to databases of protein sequences derived from the conceptual translation of genes.
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution.
The peptidyl transferase is an aminoacyltransferase as well as the primary enzymatic function of the ribosome, which forms peptide bonds between adjacent amino acids using tRNAs during the translation process of protein biosynthesis. The substrates for the peptidyl transferase reaction are two tRNA molecules, one bearing the growing peptide chain and the other bearing the amino acid that will be added to the chain. The peptidyl chain and the amino acids are attached to their respective tRNAs via ester bonds to the O atom at the CCA-3' ends of these tRNAs. Peptidyl transferase is an enzyme that catalyzes the addition of an amino acid residue in order to grow the polypeptide chain in protein synthesis. It is located in the large ribosomal subunit, where it catalyzes the peptide bond formation. It is composed entirely of RNA. The alignment between the CCA ends of the ribosome-bound peptidyl tRNA and aminoacyl tRNA in the peptidyl transferase center contribute to its ability to catalyze these reactions. This reaction occurs via nucleophilic displacement. The amino group of the aminoacyl tRNA attacks the terminal carboxyl group of the peptidyl tRNA. Peptidyl transferase activity is carried out by the ribosome. Peptidyl transferase activity is not mediated by any ribosomal proteins but by ribosomal RNA (rRNA), a ribozyme. Ribozymes are the only enzymes which are not made up of proteins, but ribonucleotides. All other enzymes are made up of proteins. This RNA relic is the most significant piece of evidence supporting the RNA World hypothesis.

Epitope mapping is the process of experimentally identifying the binding site, or "epitope", of an antibody on its target antigen. Identification and characterization of antibody binding sites aid in the discovery and development of new therapeutics, vaccines, and diagnostics. Epitope characterization can also help elucidate the mechanism of binding for an antibody and can strengthen intellectual property (patent) protection. Experimental epitope mapping data can be incorporated into robust algorithms to facilitate in silico prediction of B-cell epitopes based on sequence and/or structural data. Epitopes are generally divided into three classes: linear, conformational and discontinuous. Linear epitopes are formed by a continuous sequence of amino acids in a protein. In Conformational epitopes the binding residues are contained within certain key protein structural conformations, such as in helices, loops or beta sheets. The conformation of the epitope is important for bringing binding residues in the correct positions. Finally, discontinuous epitopes consist of parts of the antigen that are not close in the protein sequence but are brought together by three-dimensional protein folding. Discontinuous epitopes can harbour linear and conformational parts. B-cell epitope mapping studies suggest that most interactions between antigens and antibodies, particularly autoantibodies and protective antibodies, rely on binding to discontinuous epitopes.

A single-domain antibody (sdAb), also known as a nanobody, is an antibody fragment consisting of a single monomeric variable antibody domain. Like a whole antibody, it is able to bind selectively to a specific antigen. With a molecular weight of only 12–15 kDa, single-domain antibodies are much smaller than common antibodies which are composed of two heavy protein chains and two light chains, and even smaller than Fab fragments and single-chain variable fragments.
Protein tags are peptide sequences genetically grafted onto a recombinant protein. Often these tags are removable by chemical agents or by enzymatic means, such as proteolysis or intein splicing. Tags are attached to proteins for various purposes. They can be added to either end of the target protein, so they are either C-terminus or N-terminus specific or are both C-terminus and N-terminus specific. Some tags are also inserted into the coding sequence of the protein of interest; they are known as internal tags.
The Transporter Classification Database is an International Union of Biochemistry and Molecular Biology (IUBMB)-approved classification system for membrane transport proteins, including ion channels.
A peptide sequence tag is a piece of information about a peptide obtained by tandem mass spectrometry that can be used to identify this peptide in a protein database.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE) or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.
An Isotope-coded affinity tag (ICAT) is an in-vitro isotopic labeling method used for quantitative proteomics by mass spectrometry that uses chemical labeling reagents. These chemical probes consist of three elements: a reactive group for labeling an amino acid side chain, an isotopically coded linker, and a tag for the affinity isolation of labeled proteins/peptides. The samples are combined and then separated through chromatography, then sent through a mass spectrometer to determine the mass-to-charge ratio between the proteins. Only cysteine containing peptides can be analysed. Since only cysteine containing peptides are analysed, often the post translational modification is lost.
Human influenza hemagglutinin (HA) is a surface glycoprotein required for the infectivity of the human influenza virus. The HA tag is derived from the HA-molecule corresponding to amino acids 98-106. It has been extensively used as a general epitope tag in expression vectors. Many recombinant proteins have been engineered to express the HA tag, which does not appear to interfere with the bioactivity or the biodistribution of the recombinant protein. This tag facilitates the detection, isolation, and purification of the protein of interest.
Glycopeptides are peptides that contain carbohydrate moieties (glycans) covalently attached to the side chains of the amino acid residues that constitute the peptide.







