In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 of one alpha-amino acid and N2 of another, along a peptide or protein chain.

In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The part of an antibody that binds to the epitope is called a paratope. Although epitopes are usually non-self proteins, sequences derived from the host that can be recognized are also epitopes.

A ubiquitin ligase is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases regulates diverse areas such as cell trafficking, DNA repair, and signaling and is of profound importance in cell biology. E3 ligases are also key players in cell cycle control, mediating the degradation of cyclins, as well as cyclin dependent kinase inhibitor proteins. The human genome encodes over 600 putative E3 ligases, allowing for tremendous diversity in substrates.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.

In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. Protecting group strategies are usually necessary to prevent undesirable side reactions with the various amino acid side chains. Chemical peptide synthesis most commonly starts at the carboxyl end of the peptide (C-terminus), and proceeds toward the amino-terminus (N-terminus). Protein biosynthesis in living organisms occurs in the opposite direction.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.

Streptavidin is a 52 kDa protein (tetramer) purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin. With a dissociation constant (Kd) on the order of ≈10−14 mol/L, the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants, detergents, proteolytic enzymes, and extremes of temperature and pH.
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits, and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits, or two heterodimer subunits.
Protein tags are peptide sequences genetically grafted onto a recombinant protein. Tags are attached to proteins for various purposes. They can be added to either end of the target protein, so they are either C-terminus or N-terminus specific or are both C-terminus and N-terminus specific. Some tags are also inserted at sites within the protein of interest; they are known as internal tags.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.
Pilin refers to a class of fibrous proteins that are found in pilus structures in bacteria. These structures can be used for the exchange of genetic material, or as a cell adhesion mechanism. Although not all bacteria have pili or fimbriae, bacterial pathogens often use their fimbriae to attach to host cells. In Gram-negative bacteria, where pili are more common, individual pilin molecules are linked by noncovalent protein-protein interactions, while Gram-positive bacteria often have polymerized LPXTG pilin.

An isopeptide bond is a type of amide bond formed between a carboxyl group of one amino acid and an amino group of another. An isopeptide bond is the linkage between the side chain amino or carboxyl group of one amino acid to the α-carboxyl, α-amino group, or the side chain of another amino acid. In a typical peptide bond, also known as eupeptide bond, the amide bond always forms between the α-carboxyl group of one amino acid and the α-amino group of the second amino acid. Isopeptide bonds are rarer than regular peptide bonds. Isopeptide bonds lead to branching in the primary sequence of a protein. Proteins formed from normal peptide bonds typically have a linear primary sequence.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo. The consensus sequence recognized by TEV protease is Glu-Asn-Leu-Tyr-Phe-Gln-|-Ser, where "|" denotes cleaved peptide bond.
Bioconjugation is a chemical strategy to form a stable covalent link between two molecules, at least one of which is a biomolecule.
Bissulfosuccinimidyl suberate (BS3) is a crosslinker used in biological research. It is a water-soluble version of disuccinimidyl suberate.
Sortase refers to a group of prokaryotic enzymes that modify surface proteins by recognizing and cleaving a carboxyl-terminal sorting signal. For most substrates of sortase enzymes, the recognition signal consists of the motif LPXTG (Leu-Pro-any-Thr-Gly), then a highly hydrophobic transmembrane sequence, followed by a cluster of basic residues such as arginine. Cleavage occurs between the Thr and Gly, with transient attachment through the Thr residue to the active site Cys residue, followed by transpeptidation that attaches the protein covalently to cell wall components. Sortases occur in almost all Gram-positive bacteria and the occasional Gram-negative bacterium or Archaea, where cell wall LPXTG-mediated decoration has not been reported. Although sortase A, the "housekeeping" sortase, typically acts on many protein targets, other forms of sortase recognize variant forms of the cleavage motif, or catalyze the assembly of pilins into pili.
Isopeptag is a 16-amino acid peptide tag (TDKDMTITFTNKKDAE) that can be genetically linked to proteins without interfering with protein folding. What makes the isopeptag different from other peptide tags is that it can bind its binding protein through a permanent and irreversible covalent bond. Other peptide tags generally bind their targets through weak non-covalent interactions, thus limiting their use in applications where molecules experience extreme forces. The isopeptag's covalent binding to its target overcomes these barriers and allows target proteins to be studied in harsher molecular environments.
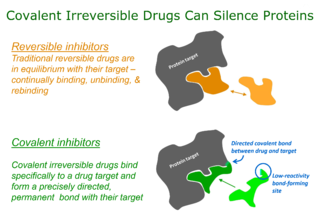
Targeted covalent inhibitors (TCIs) or Targeted covalent drugs are rationally designed inhibitors that bind and then bond to their target proteins. These inhibitors possess a bond-forming functional group of low chemical reactivity that, following binding to the target protein, is positioned to react rapidly with a proximate nucleophilic residue at the target site to form a bond.

HaloTag is a self-labeling protein tag. It is a 297 residue protein derived from a bacterial enzyme, designed to covalently bind to a synthetic ligand. The bacterial enzyme can be fused to various proteins of interest. The synthetic ligand is chosen from a number of available ligands in accordance with the type of experiments to be performed. This bacterial enzyme is a haloalkane dehalogenase, which acts as a hydrolase and is designed to facilitate visualization of the subcellular localization of a protein of interest, immobilization of a protein of interest, or capture of the binding partners of a protein of interest within its biochemical environment. The HaloTag is composed of two covalently bound segments including a haloalkane dehalogenase and a synthetic ligand of choice. These synthetic ligands consist of a reactive chloroalkane linker bound to a functional group. Functional groups can either be biotin or can be chosen from five available fluorescent dyes including Coumarin, Oregon Green, Alexa Fluor 488, diAcFAM, and TMR. These fluorescent dyes can be used in the visualization of either living or chemically fixed cells.