DNA topoisomerase 2-beta is an enzyme that in humans is encoded by the TOP2B gene. [5] [6]
DNA topoisomerase 2-beta is an enzyme that in humans is encoded by the TOP2B gene. [5] [6]
This gene encodes a DNA topoisomerase, an enzyme that controls and alters the topologic states of DNA during transcription. This nuclear enzyme is involved in processes such as chromosome condensation, chromatid separation, and the relief of torsional stress that occurs during DNA transcription and replication. It catalyzes the transient breaking and rejoining of two strands of duplex DNA which allows the strands to pass through one another, thus altering the topology of DNA. Two forms of this enzyme exist as likely products of a gene duplication event. The gene encoding this form, beta, is localized to chromosome 3 and the alpha form is localized to chromosome 17. The gene encoding this enzyme functions as the target for several anticancer agents, for example mitoxantrone, and a variety of mutations in this gene have been associated with the development of drug resistance. Reduced activity of this enzyme may also play a role in ataxia-telangiectasia. Alternative splicing of this gene results in two transcript variants; however, the second variant has not yet been fully described. [7]
During a new learning experience, a set of genes is rapidly expressed in the brain. This induced gene expression is considered to be essential for processing the information being learned. Such genes are referred to as immediate early genes (IEGs). TOP2B activity is essential for the expression of IEGs in a type of learning experience in mice termed associative fear memory. [8] Such a learning experience appears to rapidly trigger TOP2B to induce double-strand breaks in the promoter DNA of IEG genes that function in neuroplasticity. Repair of these induced breaks is associated with DNA demethylation of IEG gene promoters allowing immediate expression of these IEG genes. [8]
Activation of more than 600 regulatory sequences in promoters and 800 regulatory sequences in enhancers, in many cell types, appears to depend on short-term double-strand breaks initiated by TOP2B. [9] [10] The induction of particular double-strand breaks are specific with respect to their inducing signal. When neurons are activated in vitro, just 22 of TOP2B-induced double-strand breaks occur in their genomes, largely at immediate early genes. [11]

The induction of short-term double-strand breaks by TOP2B occurs in association with at least four enzymes of the non-homologous end joining (NHEJ) DNA repair pathway (DNA-PKcs, KU70, KU80 and DNA LIGASE IV) (see Figure). These enzymes repair the double-strand breaks within about 15 minutes to two hours. [11] [12] The double-strand breaks in the promoter are thus associated with TOP2B and at least these four repair enzymes. These proteins are present simultaneously on a single promoter nucleosome (there are about 147 nucleotides in the DNA sequence wrapped around a single nucleosome) located near the transcription start site of their target gene. [12]
The double-strand break introduced by TOP2B apparently frees a part of the promoter at an RNA polymerase-bound transcription start site to physically move to its associated enhancer (see regulatory sequence). This allows the enhancer, with its bound transcription factors and mediator proteins, to directly interact with the RNA polymerase paused at the transcription start site to start transcription. [11] [13]
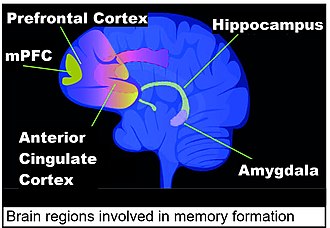
Contextual fear conditioning in the mouse causes the mouse to have a long-term memory and fear of the location in which it occurred. This conditioning causes hundreds of gene-associated DSBs in the medial prefrontal cortex (mPFC) and hippocampus that are important for learning and memory. [14]
Interactions of TOP2B and DNA also regulate transcription of genes that are important for development. These include genes encoding axon guidance factors and cell adhesion molecules. Specifically, TOP2B is required for lamina-specific targeting of retinal ganglion cell axons and dendrites in the zebrafish. [15]
TOP2B has been shown to interact with:
In Drosophila Hadlaczky et al. 1988 found DNA topoisomerase II β did not correlate with cell proliferation - while α did. [22]