The Aquificota phylum is a diverse collection of bacteria that live in harsh environmental settings. The name Aquificota was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
The ribosomal DNA consists of a group of ribosomal RNA encoding genes and related regulatory elements, and is widespread in similar configuration in all domains of life. The ribosomal DNA encodes the non-coding ribosomal RNA, integral structural elements in the assembly of ribosomes, it's importance making it the most abundant section of RNA found in cells of eukaryotes. Additionally, these segments includes regulatory sections, such as an promotor specific to the RNA polymerase I, as well as both transcribed and non-transcribed spacer segments.

The Thermodesulfobacteriota are a phylum of thermophilic sulfate-reducing bacteria.
The Thermotogota are a phylum of the domain Bacteria. The phylum contains a single class, Thermotogae. The phylum Thermotogota is composed of Gram-negative staining, anaerobic, and mostly thermophilic and hyperthermophilic bacteria.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins, though this ratio differs between prokaryotes and eukaryotes.

Sulfur-reducing bacteria are microorganisms able to reduce elemental sulfur (S0) to hydrogen sulfide (H2S). These microbes use inorganic sulfur compounds as electron acceptors to sustain several activities such as respiration, conserving energy and growth, in absence of oxygen. The final product of these processes, sulfide, has a considerable influence on the chemistry of the environment and, in addition, is used as electron donor for a large variety of microbial metabolisms. Several types of bacteria and many non-methanogenic archaea can reduce sulfur. Microbial sulfur reduction was already shown in early studies, which highlighted the first proof of S0 reduction in a vibrioid bacterium from mud, with sulfur as electron acceptor and H
2 as electron donor. The first pure cultured species of sulfur-reducing bacteria, Desulfuromonas acetoxidans, was discovered in 1976 and described by Pfennig Norbert and Biebel Hanno as an anaerobic sulfur-reducing and acetate-oxidizing bacterium, not able to reduce sulfate. Only few taxa are true sulfur-reducing bacteria, using sulfur reduction as the only or main catabolic reaction. Normally, they couple this reaction with the oxidation of acetate, succinate or other organic compounds. In general, sulfate-reducing bacteria are able to use both sulfate and elemental sulfur as electron acceptors. Thanks to its abundancy and thermodynamic stability, sulfate is the most studied electron acceptor for anaerobic respiration that involves sulfur compounds. Elemental sulfur, however, is very abundant and important, especially in deep-sea hydrothermal vents, hot springs and other extreme environments, making its isolation more difficult. Some bacteria – such as Proteus, Campylobacter, Pseudomonas and Salmonella – have the ability to reduce sulfur, but can also use oxygen and other terminal electron acceptors.

The 5S ribosomal RNA is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life, with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation coefficient in an ultracentrifuge, which is measured in Svedberg units (S).

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
Thermococcus celer is a Gram-negative, spherical-shaped archaeon of the genus Thermococcus. The discovery of T. celer played an important role in rerooting the tree of life when T. celer was found to be more closely related to methanogenic Archaea than to other phenotypically similar thermophilic species. T. celer was the first archaeon discovered to house a circularized genome. Several type strains of T. celer have been identified: Vu13, ATCC 35543, and DSM 2476.
Hydrogenobacter thermophilus is an extremely thermophilic, straight rod (bacillus) bacterium. TK-6 is the type strain for this species. It is a Gram negative, non-motile, obligate chemolithoautotroph. It belongs to one of the earliest branching order of Bacteria. H. thermophilus TK-6 lives in soil that contains hot water. It was one of the first hydrogen oxidizing bacteria described leading to the discovery, and subsequent examination of many unique proteins involved in its metabolism. Its discovery contradicted the idea that no obligate hydrogen oxidizing bacteria existed, leading to a new understanding of this physiological group. Additionally, H. thermophilus contains a fatty acid composition that had not been observed before.
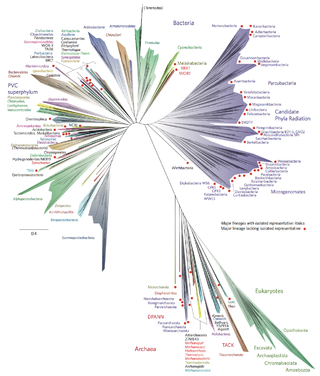
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 1987 paper by Carl Woese.
Desulfobulbus propionicus is a Gram-negative, anaerobic chemoorganotroph. Three separate strains have been identified: 1pr3T, 2pr4, and 3pr10. It is also the first pure culture example of successful disproportionation of elemental sulfur to sulfate and sulfide. Desulfobulbus propionicus has the potential to produce free energy and chemical products.
Sulfolobus metallicus is a coccoid shaped thermophilic archaeon. It is a strict chemolithoautotroph gaining energy by oxidation of sulphur and sulphidic ores into sulfuric acid. Its type strain is Kra 23. It has many uses that take advantage of its ability to grow on metal media under acidic and hot environments.
Acidithiobacillus caldus formerly belonged to the genus Thiobacillus prior to 2000, when it was reclassified along with a number of other bacterial species into one of three new genera that better categorize sulfur-oxidizing acidophiles. As a member of the Gammaproteobacteria class of Pseudomonadota, A. caldus may be identified as a Gram-negative bacterium that is frequently found in pairs. Considered to be one of the most common microbes involved in biomining, it is capable of oxidizing reduced inorganic sulfur compounds (RISCs) that form during the breakdown of sulfide minerals. The meaning of the prefix acidi- in the name Acidithiobacillus comes from the Latin word acidus, signifying that members of this genus love a sour, acidic environment. Thio is derived from the Greek word thios and describes the use of sulfur as an energy source, and bacillus describes the shape of these microorganisms, which are small rods. The species name, caldus, is derived from the Latin word for warm or hot, denoting this species' love of a warm environment.
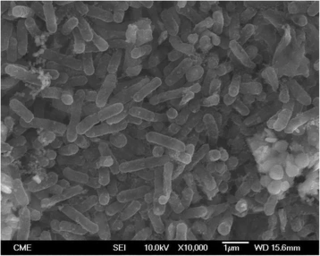
Acidithiobacillus thiooxidans, formerly known as Thiobacillus thiooxidans until its reclassification into the newly designated genus Acidithiobacillus of the Acidithiobacillia subclass of Pseudomonadota, is a Gram-negative, rod-shaped bacterium that uses sulfur as its primary energy source. It is mesophilic, with a temperature optimum of 28 °C. This bacterium is commonly found in soil, sewer pipes, and cave biofilms called snottites. A. thiooxidans is used in the mining technique known as bioleaching, where metals are extracted from their ores through the action of microbes.
Geobacillus jurassicus is a thermophilic bacterium first isolated from a high-temperature petroleum reservoir. It is aerobic, gram-positive, rod-shaped, moderately thermophilic, chemoorganotrophic, and endospore-forming, with type species DS1T.
Desulfovibrio alcoholivorans is a bacterium from the genus of Desulfovibrio which has been isolated from alcohol industry waste water in France.
Thiodictyon is a genus of gram-negative bacterium classified within purple sulfur bacteria (PSB).
Fervidicoccus fontis is an extremophilic, coccus-shaped archaeaon known for thriving in high-temperature environments. It was discovered in Russia's Uzon Caldera and exhibits anaerobic, organotrophic metabolism, primarily fermenting organic compounds such as peptides and yeast extract. F. fontis is genetically distinct, sharing no more than 89% of its genetic material with its closest relatives. It is the sole species within the order Fervidicoccales and genus Fervidicoccus, although ongoing research suggests potential new species. It plays a significant role in biotechnological applications due to its lipid-hydrolyzing capabilities, contributing to industries ranging from wastewater treatment to pharmaceuticals.






