
Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual mode of reproduction in bacteria.

A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet.
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid, used for transforming and cloning in bacteria, usually E. coli. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150–350 kbp. A similar cloning vector called a PAC has also been produced from the DNA of P1 bacteriophage.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

Stanley Norman Cohen is an American geneticist and the Kwoh-Ting Li Professor in the Stanford University School of Medicine. Stanley Cohen and Herbert Boyer were the first scientists to transplant genes from one living organism to another, a fundamental discovery for genetical engineering. Thousands of products have been developed on the basis of their work, including human growth hormone and hepatitis B vaccine. According to immunologist Hugh McDevitt, "Cohen's DNA cloning technology has helped biologists in virtually every field". Without it, "the face of biomedicine and biotechnology would look totally different." Boyer cofounded Genentech in 1976 based on their work together, but Cohen was a consultant for Cetus Corporation and declined to join. In 2022, Cohen was found guilty of having committed fraud in misleading investors into a biotechnology company he founded in 2016, and paid $29 million in damages.

A plasmid preparation is a method of DNA extraction and purification for plasmid DNA, it is an important step in many molecular biology experiments and is essential for the successful use of plasmids in research and biotechnology. Many methods have been developed to purify plasmid DNA from bacteria. During the purification procedure, the plasmid DNA is often separated from contaminating proteins and genomic DNA.
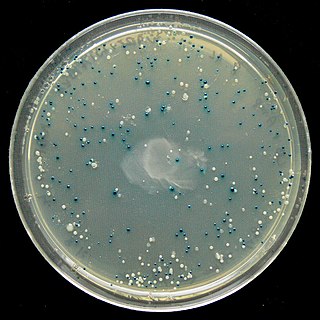
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used. DNA of transformation is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.
An origin of transfer (oriT) is a short sequence ranging from 40-500 base pairs in length that is necessary for the transfer of DNA from a gram-negative bacterial donor to recipient during bacterial conjugation. The transfer of DNA is a critical component for antimicrobial resistance within bacterial cells and the oriT structure and mechanism within plasmid DNA is complementary to its function in bacterial conjugation. The first oriT to be identified and cloned was on the RK2 (IncP) conjugative plasmid, which was done by Guiney and Helinski in 1979.

Functional cloning is a molecular cloning technique that relies on prior knowledge of the encoded protein’s sequence or function for gene identification. In this assay, a genomic or cDNA library is screened to identify the genetic sequence of a protein of interest. Expression cDNA libraries may be screened with antibodies specific for the protein of interest or may rely on selection via the protein function. Historically, the amino acid sequence of a protein was used to prepare degenerate oligonucleotides which were then probed against the library to identify the gene encoding the protein of interest. Once candidate clones carrying the gene of interest are identified, they are sequenced and their identity is confirmed. This method of cloning allows researchers to screen entire genomes without prior knowledge of the location of the gene or the genetic sequence.
A P1-derived artificial chromosome, or PAC, is a DNA construct derived from the DNA of P1 bacteriophages and Bacterial artificial chromosome. It can carry large amounts of other sequences for a variety of bioengineering purposes in bacteria. It is one type of the efficient cloning vector used to clone DNA fragments in Escherichia coli cells.
In molecular cloning, a vector is any particle used as a vehicle to artificially carry a foreign nucleic sequence – usually DNA – into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors are an origin of replication, a multicloning site, and a selectable marker.

The E. coli long-term evolution experiment (LTEE) is an ongoing study in experimental evolution begun by Richard Lenski at the University of California, Irvine, carried on by Lenski and colleagues at Michigan State University, and currently overseen by Jeffrey E. Barrick at the University of Texas at Austin. It has been tracking genetic changes in 12 initially identical populations of asexual Escherichia coli bacteria since 24 February 1988. Lenski performed the 10,000th transfer of the experiment on March 13, 2017. The populations reached over 73,000 generations in early 2020, shortly before being frozen because of the COVID-19 pandemic. In September 2020, the LTEE experiment was resumed using the frozen stocks.
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.

Thermotoga maritima is a hyperthermophilic, anaerobic organism that is a member of the order Thermotogales. T. maritima is well known for its ability to produce hydrogen (clean energy) and it is the only fermentative bacterium that has been shown to produce Hydrogen more than the Thauer limit (>4 mol H2 /mol glucose). It employs [FeFe]-hydrogenases to produce hydrogen gas (H2) by fermenting many different types of carbohydrates.
Bacterial genomes are generally smaller and less variant in size among species when compared with genomes of eukaryotes. Bacterial genomes can range in size anywhere from about 130 kbp to over 14 Mbp. A study that included, but was not limited to, 478 bacterial genomes, concluded that as genome size increases, the number of genes increases at a disproportionately slower rate in eukaryotes than in non-eukaryotes. Thus, the proportion of non-coding DNA goes up with genome size more quickly in non-bacteria than in bacteria. This is consistent with the fact that most eukaryotic nuclear DNA is non-gene coding, while the majority of prokaryotic, viral, and organellar genes are coding. Right now, we have genome sequences from 50 different bacterial phyla and 11 different archaeal phyla. Second-generation sequencing has yielded many draft genomes ; third-generation sequencing might eventually yield a complete genome in a few hours. The genome sequences reveal much diversity in bacteria. Analysis of over 2000 Escherichia coli genomes reveals an E. coli core genome of about 3100 gene families and a total of about 89,000 different gene families. Genome sequences show that parasitic bacteria have 500–1200 genes, free-living bacteria have 1500–7500 genes, and archaea have 1500–2700 genes. A striking discovery by Cole et al. described massive amounts of gene decay when comparing Leprosy bacillus to ancestral bacteria. Studies have since shown that several bacteria have smaller genome sizes than their ancestors did. Over the years, researchers have proposed several theories to explain the general trend of bacterial genome decay and the relatively small size of bacterial genomes. Compelling evidence indicates that the apparent degradation of bacterial genomes is owed to a deletional bias.
Thermotoga naphthophila is a hyperthermophilic, anaerobic, non-spore-forming, rod-shaped fermentative heterotroph, with type strain RKU-10T.
Fervidobacterium islandicum is a species of extremely thermophilic anaerobic bacteria, first isolated from an Icelandic hot spring.
Bacterial recombination is a type of genetic recombination in bacteria characterized by DNA transfer from one organism called donor to another organism as recipient. This process occurs in three main ways:










