Mutagenesis is a process by which the genetic information of an organism is changed by the production of a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using laboratory procedures. A mutagen is a mutation-causing agent, be it chemical or physical, which results in an increased rate of mutations in an organism's genetic code. In nature mutagenesis can lead to cancer and various heritable diseases, and it is also a driving force of evolution. Mutagenesis as a science was developed based on work done by Hermann Muller, Charlotte Auerbach and J. M. Robson in the first half of the 20th century.
Genotoxicity is the property of chemical agents that damage the genetic information within a cell causing mutations, which may lead to cancer. While genotoxicity is often confused with mutagenicity, all mutagens are genotoxic, but some genotoxic substances are not mutagenic. The alteration can have direct or indirect effects on the DNA: the induction of mutations, mistimed event activation, and direct DNA damage leading to mutations. The permanent, heritable changes can affect either somatic cells of the organism or germ cells to be passed on to future generations. Cells prevent expression of the genotoxic mutation by either DNA repair or apoptosis; however, the damage may not always be fixed leading to mutagenesis.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encodes its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.

A molecular lesion or point lesion is damage to the structure of a biological molecule such as DNA, RNA, or protein. This damage may result in the reduction or absence of normal function, and in rare cases the gain of a new function. Lesions in DNA may consist of breaks or other changes in chemical structure of the helix, ultimately preventing transcription. Meanwhile, lesions in proteins consist of both broken bonds and improper folding of the amino acid chain. While many nucleic acid lesions are general across DNA and RNA, some are specific to one, such as thymine dimers being found exclusively in DNA. Several cellular repair mechanisms exist, ranging from global to specific, in order to prevent lasting damage resulting from lesions.

Benzo[a]pyrene (BaP or B[a]P) is a polycyclic aromatic hydrocarbon and the result of incomplete combustion of organic matter at temperatures between 300 °C (572 °F) and 600 °C (1,112 °F). The ubiquitous compound can be found in coal tar, tobacco smoke and many foods, especially grilled meats. The substance with the formula C20H12 is one of the benzopyrenes, formed by a benzene ring fused to pyrene. Its diol epoxide metabolites, more commonly known as BPDE, react with and bind to DNA, resulting in mutations and eventually cancer. It is listed as a Group 1 carcinogen by the IARC. In the 18th century a scrotal cancer of chimney sweepers, the chimney sweeps' carcinoma, was already known to be connected to soot.

A germline mutation, or germinal mutation, is any detectable variation within germ cells. Mutations in these cells are the only mutations that can be passed on to offspring, when either a mutated sperm or oocyte come together to form a zygote. After this fertilization event occurs, germ cells divide rapidly to produce all of the cells in the body, causing this mutation to be present in every somatic and germline cell in the offspring; this is also known as a constitutional mutation. Germline mutation is distinct from somatic mutation.
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.

Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.

MUTYH is a human gene that encodes a DNA glycosylase, MUTYH glycosylase. It is involved in oxidative DNA damage repair and is part of the base excision repair pathway. The enzyme excises adenine bases from the DNA backbone at sites where adenine is inappropriately paired with guanine, cytosine, or 8-oxo-7,8-dihydroguanine, a common form of oxidative DNA damage.
DNA oxidation is the process of oxidative damage of deoxyribonucleic acid. As described in detail by Burrows et al., 8-oxo-2'-deoxyguanosine (8-oxo-dG) is the most common oxidative lesion observed in duplex DNA because guanine has a lower one-electron reduction potential than the other nucleosides in DNA. The one electron reduction potentials of the nucleosides are guanine 1.29, adenine 1.42, cytosine 1.6 and thymine 1.7. About 1 in 40,000 guanines in the genome are present as 8-oxo-dG under normal conditions. This means that >30,000 8-oxo-dGs may exist at any given time in the genome of a human cell. Another product of DNA oxidation is 8-oxo-dA. 8-oxo-dA occurs at about 1/10 the frequency of 8-oxo-dG. The reduction potential of guanine may be reduced by as much as 50%, depending on the particular neighboring nucleosides stacked next to it within DNA.
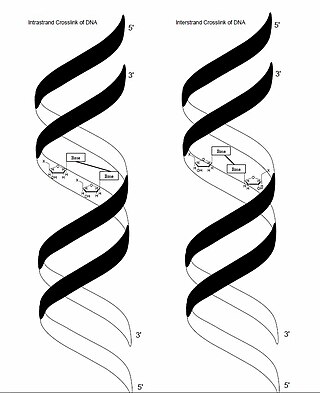
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.

8-Oxoguanine glycosylase, also known as OGG1, is a DNA glycosylase enzyme that, in humans, is encoded by the OGG1 gene. It is involved in base excision repair. It is found in bacterial, archaeal and eukaryotic species.

8-Oxoguanine (8-hydroxyguanine, 8-oxo-Gua, or OH8Gua) is one of the most common DNA lesions resulting from reactive oxygen species modifying guanine, and can result in a mismatched pairing with adenine resulting in G to T and C to A substitutions in the genome. In humans, it is primarily repaired by DNA glycosylase OGG1. It can be caused by ionizing radiation, in connection with oxidative metabolism.

8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress.
DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a nucleobase missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors, but is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage.
DNA polymerase IV is a prokaryotic polymerase that is involved in mutagenesis and is encoded by the dinB gene. It exhibits no 3′→5′ exonuclease (proofreading) activity and hence is error prone. In E. coli, DNA polymerase IV is involved in non-targeted mutagenesis. Pol IV is a Family Y polymerase expressed by the dinB gene that is switched on via SOS induction caused by stalled polymerases at the replication fork. During SOS induction, Pol IV production is increased tenfold and one of the functions during this time is to interfere with Pol III holoenzyme processivity. This creates a checkpoint, stops replication, and allows time to repair DNA lesions via the appropriate repair pathway. Another function of Pol IV is to perform translesion synthesis at the stalled replication fork like, for example, bypassing N2-deoxyguanine adducts at a faster rate than transversing undamaged DNA. Cells lacking dinB gene have a higher rate of mutagenesis caused by DNA damaging agents.
2-hydroxy-dATP diphosphatase is an enzyme that in humans is encoded by the NUDT1 gene. During DNA repair, the enzyme hydrolyses oxidized purines and prevents their addition onto the DNA chain. As such it has important role in aging and cancer development.
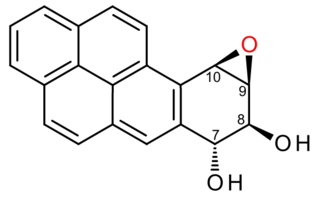
(+)-Benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide is an organic compound with molecular formula C20H14O3. It is a metabolite and derivative of benzo[a]pyrene (found in tobacco smoke) as a result of oxidation to include hydroxyl and epoxide functionalities. (+)-Benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide binds to the N2 atom of a guanine nucleobase in DNA, distorting the double helix structure by intercalation of the pyrene moiety between base pairs through π-stacking. The carcinogenic properties of tobacco smoking are attributed in part to this compound binding and inactivating the tumor suppression ability of certain genes, leading to genetic mutations and potentially to cancer.

Cynthia J. Burrows is an American chemist, currently a distinguished professor in the department of chemistry at the University of Utah, where she is also the Thatcher Presidential Endowed Chair of Biological Chemistry. Burrows was the Senior Editor of the Journal of Organic Chemistry (2001-2013) and became Editor-in-Chief of Accounts of Chemical Research in 2014.,,
Mutational signatures are characteristic combinations of mutation types arising from specific mutagenesis processes such as DNA replication infidelity, exogenous and endogenous genotoxin exposures, defective DNA repair pathways, and DNA enzymatic editing.












