
Mycoplasma is a genus of bacteria that, like the other members of the class Mollicutes, lack a cell wall around their cell membranes. Peptidoglycan (murein) is absent. This characteristic makes them naturally resistant to antibiotics that target cell wall synthesis. They can be parasitic or saprotrophic. Several species are pathogenic in humans, including M. pneumoniae, which is an important cause of "walking" pneumonia and other respiratory disorders, and M. genitalium, which is believed to be involved in pelvic inflammatory diseases. Mycoplasma species are among the smallest organisms yet discovered, can survive without oxygen, and come in various shapes. For example, M. genitalium is flask-shaped, while M. pneumoniae is more elongated, many Mycoplasma species are coccoid. Hundreds of Mycoplasma species infect animals.

Campylobacter jejuni is a species of pathogenic bacteria, one of the most common causes of food poisoning in Europe and in the US. The vast majority of cases occur as isolated events, not as part of recognized outbreaks. Active surveillance through the Foodborne Diseases Active Surveillance Network (FoodNet) indicates that about 20 cases are diagnosed each year for each 100,000 people in the US, while many more cases are undiagnosed or unreported; the CDC estimates a total of 1.5 million infections every year. The European Food Safety Authority reported 246,571 cases in 2018, and estimated approximately nine million cases of human campylobacteriosis per year in the European Union.
Ureaplasma urealyticum is a bacterium belonging to the genus Ureaplasma and the family Mycoplasmataceae in the order Mycoplasmatales. This family consists of the genera Mycoplasma and Ureaplasma. Its type strain is T960. There are two known biovars of this species; T960 and 27. These strains of bacteria are commonly found as commensals in the urogenital tracts of human beings, but overgrowth can lead to infections that cause the patient discomfort. Unlike most bacteria, Ureaplasma urealyticum lacks a cell wall making it unique in physiology and medical treatment.
Mycoplasmataceae is a family of bacteria in the order Mycoplasmatales. This family consists of the genera Mycoplasma and Ureaplasma.

Campylobacterota are a phylum of bacteria. All species of this phylum are Gram-negative.

Ehrlichia ewingii is a species of rickettsiales bacteria. It has recently been associated with human infection, and can be detected via PCR serological testing. The name Ehrlichia ewingii was proposed in 1992.
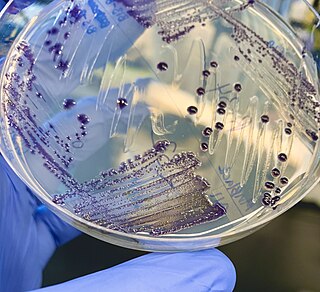
The genus Massilia belongs to the family Oxalobacteriaceae, and describes a group of gram-negative, motile, rod-shaped cells. They may contain either peritrichous or polar flagella. This genus was first described in 1998, after the type species, Massilia timonae, was isolated from the blood of an immunocompromised patient. The genus was named after the old Greek and Roman name for the city of Marseille, France, where the organism was first isolated. The Massilia genus is a diverse group that resides in many different environments, has many heterotrophic means of gathering energy, and is commonly found in association with plants.
Afipia birgiae is a species in the Afipia bacterial genus. It is a gram-negative, oxidase-positive rod in the alpha-2 subgroup of the class Proteobacteria. It is motile by means of a single flagellum. Its type strain is 34632T.
Brucella microti is a species of bacteria first isolated from the common vole, Microtus arvalis. Its genome has been sequenced. It is Gram-negative, non-motile, non-spore-forming, and coccoid, with the type strain CCM 4915T. It is pathogenic.
Ureaplasma canigenitalium is a species of Ureaplasma, a genus of bacteria belonging to the family Mycoplasmataceae. It has been isolated from dogs. It possesses the sequence accession no. for the type strain: D78648.
Ureaplasma felinum is a species of Ureaplasma, a genus of bacteria belonging to the family Mycoplasmataceae. It has been isolated from cats. It possesses the sequence accession no. for the type strain: D78651.
Mycoplasma orale is a small bacterium found in the class Mollicutes. It belongs to the genus Mycoplasma, a well-known group of bacterial parasites that inhabit humans. It also is known to be an opportunistic pathogen in immunocompromised humans. As with other Mycoplasma species, M. orale is not readily treated with many antibiotics due to its lack of a peptidoglycan cell wall. Therefore, this species is relevant to the medical field as physicians face the task of treating patients infected with this microbe. It is characterized by a small physical size, a small genome size, and a limited metabolism. It is also known to frequently contaminate laboratory experiments. This bacteria is very similar physiologically and morphologically to its sister species within the genus Mycoplasma; however, its recent discovery leaves many questions still unanswered about this microbe.
Azospirillum is a Gram-negative, microaerophilic, non-fermentative and nitrogen-fixing bacterial genus from the family of Rhodospirillaceae. Azospirillum bacteria can promote plant growth.

Phytobacter is a genus of Gram-negative bacteria emerging from the grouping of isolates previously assigned to various genera of the family Enterobacteriaceae. This genus was first established on the basis of nitrogen fixing isolates from wild rice in China, but also includes a number of isolates obtained during a 2013 multi-state sepsis outbreak in Brazil and, retrospectively, several clinical strains isolated in the 1970s in the United States that are still available in culture collections, which originally were grouped into Brenner's Biotype XII of the Erwinia herbicola-Enterobacter agglomerans-Complex (EEC). Standard biochemical evaluation panels are lacking Phytobacter spp. from their database, thus often leading to misidentifications with other Enterobacterales species, especially Pantoea agglomerans. Clinical isolates of the species have been identified as an important source of extended-spectrum β-lactamase and carbapenem-resistance genes, which are usually mediated by genetic mobile elements. Strong protection of co-infecting sensitive bacteria has also been reported. Bacteria belonging to this genus are not pigmented, chemoorganotrophic and able to fix nitrogen. They are lactose fermenting, cytochrome-oxidase negative and catalase positive. Glucose is fermented with the production of gas. Colonies growing on MacConkey agar (MAC) are circular, convex and smooth with non-entire margins and a usually elevated center. Three species are currently validly included in the genus Phytobacter, which is still included within the Kosakonia clade in the lately reviewed family of Enterobacteriaceae. The incorporation of a fourth species, Phytobacter massiliensis, has recently been proposed via the unification of the genera Metakosakonia and Phytobacter.
Endozoicomonas is a genus of Gram-negative, aerobic or facultatively anaerobic, chemoorganotrophic, rod-shaped, marine bacteria from the family of Hahellaceae. Endozoicomonas are symbionts of marine animals.
Scandinavium is a genus of Gram-negative, facultative anaerobic, oxidase-negative, rod-shaped, motile bacteria of the family Enterobacteriaceae. It contains a single species, Scandinavium goeteborgense. The type strain of the species is S. goeteborgenseCCUG 66741T = CECT 9823T = NCTC 14286T and its genome sequence is publicly available in DNA Data Bank of Japan, European Nucleotide Archive and GenBank under the accession number LYLP00000000.
Acinetobacter guerrae is a species of Gram-negative bacteria of the genus Acinetobacter that was described in 2020. The species was described based on the characterization of two strains, isolated from raw chicken meat, in Porto, Portugal. Additionally, two publicly available draft genome sequences were also identified as members of A. guerrae, one of them isolated from human sputum in Kanagawa, Japan, and one isolated from hospital sewage in Sichuan, China. The draft genome sequence of the type strain is deposited in DNA Data Bank of Japan, European Nucleotide Archive, and GenBank under the accession number LXGN00000000
Acinetobacter portensis is a species of Gram-negative bacteria of the genus Acinetobacter described in 2020. The description was based on the characterization of four strains, isolated from raw beef, chicken, pork and turkey meat, collected from supermarkets in Porto, Portugal, in 2014. The type strain of the species is available at the Culture Collection University of Gothenburg (CCUG) and the Czech Collection of Microorganisms (CCM), under the deposit numbers CCUG 68672T and CCM 8789T, respectively. The draft genome sequence of the type strain is deposited in DNA Data Bank of Japan, European Nucleotide Archive and GenBank under the accession number LWRV00000000.
Cytophagales is an order of non-spore forming, rod-shaped, Gram-negative bacteria that move through a gliding or flexing motion. These chemoorganotrophs are important remineralizers of organic materials into micronutrients. They are widely dispersed in the environment, found in ecosystems including soil, freshwater, seawater and sea ice. Cytophagales is included in the Bacteroidota phylum.
Neobacillus is a genus of rod-shaped bacteria that show Gram-positive or Gram-variable staining. This genus belongs under the family Bacillaceae within the order Bacillales. The type species of Neobacillus is Neobacillus niacini.




