Related Research Articles

Pathology is the study of the causes and effects of disease or injury. The word pathology also refers to the study of disease in general, incorporating a wide range of biology research fields and medical practices. However, when used in the context of modern medical treatment, the term is often used in a narrower fashion to refer to processes and tests that fall within the contemporary medical field of "general pathology", an area which includes a number of distinct but inter-related medical specialties that diagnose disease, mostly through analysis of tissue and human cell samples. Idiomatically, "a pathology" may also refer to the predicted or actual progression of particular diseases, and the affix pathy is sometimes used to indicate a state of disease in cases of both physical ailment and psychological conditions. A physician practicing pathology is called a pathologist.

A picture archiving and communication system (PACS) is a medical imaging technology which provides economical storage and convenient access to images from multiple modalities. Electronic images and reports are transmitted digitally via PACS; this eliminates the need to manually file, retrieve, or transport film jackets, the folders used to store and protect X-ray film. The universal format for PACS image storage and transfer is DICOM. Non-image data, such as scanned documents, may be incorporated using consumer industry standard formats like PDF, once encapsulated in DICOM. A PACS consists of four major components: The imaging modalities such as X-ray plain film (PF), computed tomography (CT) and magnetic resonance imaging (MRI), a secured network for the transmission of patient information, workstations for interpreting and reviewing images, and archives for the storage and retrieval of images and reports. Combined with available and emerging web technology, PACS has the ability to deliver timely and efficient access to images, interpretations, and related data. PACS reduces the physical and time barriers associated with traditional film-based image retrieval, distribution, and display.
Digital Imaging and Communications in Medicine (DICOM) is the standard for the communication and management of medical imaging information and related data. DICOM is most commonly used for storing and transmitting medical images enabling the integration of medical imaging devices such as scanners, servers, workstations, printers, network hardware, and picture archiving and communication systems (PACS) from multiple manufacturers. It has been widely adopted by hospitals and is making inroads into smaller applications such as dentists' and doctors' offices.

Medical imaging is the technique and process of imaging the interior of a body for clinical analysis and medical intervention, as well as visual representation of the function of some organs or tissues (physiology). Medical imaging seeks to reveal internal structures hidden by the skin and bones, as well as to diagnose and treat disease. Medical imaging also establishes a database of normal anatomy and physiology to make it possible to identify abnormalities. Although imaging of removed organs and tissues can be performed for medical reasons, such procedures are usually considered part of pathology instead of medical imaging.

OsiriX is an image processing application for the Apple MacOS operating system dedicated to DICOM images produced by equipment. OsiriX is complementary to existing viewers, in particular to nuclear medicine viewers. It can also read many other file formats: TIFF, JPEG, PDF, AVI, MPEG and QuickTime. It is fully compliant with the DICOM standard for image communication and image file formats. OsiriX is able to receive images transferred by DICOM communication protocol from any PACS or medical imaging modality.

Materialise Mimics is an image processing software for 3D design and modeling, developed by Materialise NV, a Belgian company specialized in additive manufacturing software and technology for medical, dental and additive manufacturing industries. Materialise Mimics is used to create 3D surface models from stacks of 2D image data. These 3D models can then be used for a variety of engineering applications. Mimics is an acronym for Materialise Interactive Medical Image Control System. It is developed in an ISO environment with CE and FDA 510k premarket clearance. Materialise Mimics is commercially available as part of the Materialise Mimics Innovation Suite, which also contains Materialise3-matic, a design and meshing software for anatomical data. The current version is 24.0(released in 2021), it supports Windows 10, Windows 7, Vista and XP in x64.
Virtual microscopy is a method of posting microscope images on, and transmitting them over, computer networks. This allows independent viewing of images by large numbers of people in diverse locations. It involves a synthesis of microscopy technologies and digital technologies. The use of virtual microscopes can transform traditional teaching methods by removing the reliance on physical space, equipment, and specimens to a model that is solely dependent upon computer-internet access. This increases the convenience of accessing the slide sets and making the slides available to a broader audience. Digitized slides can have a high resolution and are resistant to being damaged or broken over time.
Imaging informatics, also known as radiology informatics or medical imaging informatics, is a subspecialty of biomedical informatics that aims to improve the efficiency, accuracy, usability and reliability of medical imaging services within the healthcare enterprise. It is devoted to the study of how information about and contained within medical images is retrieved, analyzed, enhanced, and exchanged throughout the medical enterprise.

Digital pathology is a sub-field of pathology that focuses on data management based on information generated from digitized specimen slides. Through the use of computer-based technology, digital pathology utilizes virtual microscopy. Glass slides are converted into digital slides that can be viewed, managed, shared and analyzed on a computer monitor. With the practice of Whole-Slide Imaging (WSI), which is another name for virtual microscopy, the field of digital pathology is growing and has applications in diagnostic medicine, with the goal of achieving efficient and cheaper diagnoses, prognosis, and prediction of diseases due to the success in machine learning and artificial intelligence in healthcare.

Automated tissue image analysis or histopathology image analysis (HIMA) is a process by which computer-controlled automatic test equipment is used to evaluate tissue samples, using computations to derive quantitative measurements from an image to avoid subjective errors.

Visual artifacts are anomalies apparent during visual representation as in digital graphics and other forms of imagery, especially photography and microscopy.
Telepathology is the practice of pathology at a distance. It uses telecommunications technology to facilitate the transfer of image-rich pathology data between distant locations for the purposes of diagnosis, education, and research. Performance of telepathology requires that a pathologist selects the video images for analysis and the rendering of diagnoses. The use of "television microscopy", the forerunner of telepathology, did not require that a pathologist have physical or virtual "hands-on" involvement in the selection of microscopic fields-of-view for analysis and diagnosis.
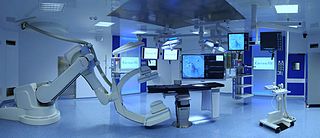
A hybrid operating room is a surgical theatre that is equipped with advanced medical imaging devices such as fixed C-Arms, X-ray computed tomography (CT) scanners or magnetic resonance imaging (MRI) scanners. These imaging devices enable minimally-invasive surgery. Minimally-invasive surgery is intended to be less traumatic for the patient and minimize incisions on the patient and perform surgery procedure through one or several small cuts.
Endomicroscopy is a technique for obtaining histology-like images from inside the human body in real-time, a process known as ‘optical biopsy’. It generally refers to fluorescence confocal microscopy, although multi-photon microscopy and optical coherence tomography have also been adapted for endoscopic use. Commercially available clinical and pre-clinical endomicroscopes can achieve a resolution on the order of a micrometre, have a field-of-view of several hundred µm, and are compatible with fluorophores which are excitable using 488 nm laser light. The main clinical applications are currently in imaging of the tumour margins of the brain and gastro-intestinal tract, particularly for the diagnosis and characterisation of Barrett’s Esophagus, pancreatic cysts and colorectal lesions. A number of pre-clinical and transnational applications have been developed for endomicroscopy as it enables researchers to perform live animal imaging. Major pre-clinical applications are in gastro-intestinal tract, toumour margin detection, uterine complications, ischaemia, live imaging of cartilage and tendon and organoid imaging.
A Vendor Neutral Archive (VNA) is a medical imaging technology in which images and documents are stored (archived) in a standard format with a standard interface, such that they can be accessed in a vendor-neutral manner by other systems.
A digital autopsy is a non-invasive autopsy in which digital imaging technology, such as with Computerized Tomography (CT) or Magnetic Resonance Imaging (MRI) scans, is used to develop three-dimensional images for a virtual exploration of a human body.
The Cancer Imaging Archive (TCIA) is an open-access database of medical images for cancer research. The site is funded by the National Cancer Institute's (NCI) Cancer Imaging Program, and the contract is operated by the University of Arkansas for Medical Sciences. Data within the archive is organized into collections which typically share a common cancer type and/or anatomical site. The majority of the data consists of CT, MRI, and nuclear medicine images stored in DICOM format, but many other types of supporting data are also provided or linked to, in order to enhance research utility. All data are de-identified in order to comply with the Health Insurance Portability and Accountability Act and National Institutes of Health data sharing policies.
Enterprise imaging has been defined as "a set of strategies, initiatives, and workflows implemented across a healthcare enterprise to consistently and optimally capture, index, manage, store, distribute, view, exchange, and analyze all clinical imaging and multimedia content to enhance the electronic health record". The concepts of enterprise imaging are elucidated in a series of papers by members of the HIMSS-SIIM Enterprise Imaging Workgroup.

Studierfenster or StudierFenster (SF) is a free, non-commercial open science client/server-based medical imaging processing online framework. It offers capabilities, like viewing medical data (computed tomography (CT), magnetic resonance imaging (MRI), etc.) in two- and three-dimensional space directly in standard web browsers, like Google Chrome, Mozilla Firefox, Safari, and Microsoft Edge. Other functionalities are the calculation of medical metrics (dice score and Hausdorff distance), manual slice-by-slice outlining of structures in medical images (segmentation), manual placing of (anatomical) landmarks in medical image data, viewing medical data in virtual reality, a facial reconstruction and registration of medical data for augmented reality, one click showcases for COVID-19 and veterinary scans, and a Radiomics module.
The Visible Embryo Project (VEP) is a multi-institutional, multidisciplinary research project originally created in the early 1990s as a collaboration between the Developmental Anatomy Center at the National Museum of Health and Medicine and the Biomedical Visualization Laboratory (BVL) at the University of Illinois at Chicago, "to develop software strategies for the development of distributed biostructural databases using cutting-edge technologies for high-performance computing and communications (HPCC), and to implement these tools in the creation of a large-scale digital archive of multidimensional data on normal and abnormal human development." This project related to BVL's other research in the areas of health informatics, educational multimedia, and biomedical imaging science. Over the following decades, the list of VEP collaborators grew to include over a dozen universities, national laboratories, and companies around the world.
References
- ↑ DICOM Standards Committee (1993). "PS3.3 - Information Object Definitions" (PDF). Retrieved 2017-10-28.
- ↑ DICOM Standards Committee (1993). "PS3.5 - Data Structures and Encoding" (PDF). Retrieved 2017-10-28.
- ↑ Bidgood, WD; Korman, LY; Golichowski, AM; Hildebrand, PL; Mori, AR; Bray, B (1997). "Controlled terminology for clinically-relevant indexing and selective retrieval of biomedical images". Int J Digit Libr. 1 (3): 278–87. doi:10.1007/s007990050022. S2CID 32595860.
- ↑ Bidgood, WD; Horii, SC (1996). "Modular extension of the ACR-NEMA DICOM standard to support new diagnostic imaging modalities and services". Journal of Digital Imaging. 9 (2): 67–77. doi: 10.1007/BF03168859 . PMID 8734576.
- ↑ Bidgood, WD; Horii, SC; Prior, FW; Van Syckle, DE (1997-01-16). "Understanding and Using DICOM, the Data Interchange Standard for Biomedical Imaging". Journal of the American Medical Informatics Association. 4 (3): 199–212. doi:10.1136/jamia.1997.0040199. PMC 61235 . PMID 9147339.
- ↑ DICOM Standards Committee (1999-07-02). "Supplement 15 - Visible Light Image Object" (PDF). Retrieved 2017-10-28.
- ↑ Dayhoff, RE (1993-06-23). "A Multidepartmental Hospital Imaging System: Implications for the Electronic Medical Record". Proceedings. The Third International Conference on Image Management and Communication in Patient Care. IMAC 93 Proceedings the Third International Conference on Image Management and Communication in Patient Care. pp. 83–6. doi:10.1109/IMAC.1993.665436. ISBN 978-0-8186-3640-0. S2CID 54209447.
- ↑ Clunie, DA; Dennison, DK; Cram, D; Persons, KR; Bronkalla, MD; Primo, H (2016). "Technical Challenges of Enterprise Imaging: HIMSS-SIIM Collaborative White Paper". Journal of Digital Imaging. 29 (5): 583–614. doi:10.1007/s10278-016-9899-4. PMC 5023533 . PMID 27576909.