
Yersinia pestis is a gram-negative, non-motile, coccobacillus bacterium without spores that is related to both Yersinia pseudotuberculosis and Yersinia enterocolitica. It is a facultative anaerobic organism that can infect humans via the Oriental rat flea. It causes the disease plague, which caused the first plague pandemic and the Black Death, the deadliest pandemic in recorded history. Plague takes three main forms: pneumonic, septicemic, and bubonic.
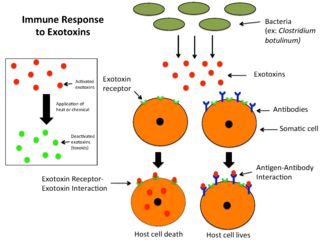
An exotoxin is a toxin secreted by bacteria. An exotoxin can cause damage to the host by destroying cells or disrupting normal cellular metabolism. They are highly potent and can cause major damage to the host. Exotoxins may be secreted, or, similar to endotoxins, may be released during lysis of the cell. Gram negative pathogens may secrete outer membrane vesicles containing lipopolysaccharide endotoxin and some virulence proteins in the bounding membrane along with some other toxins as intra-vesicular contents, thus adding a previously unforeseen dimension to the well-known eukaryote process of membrane vesicle trafficking, which is quite active at the host-pathogen interface.
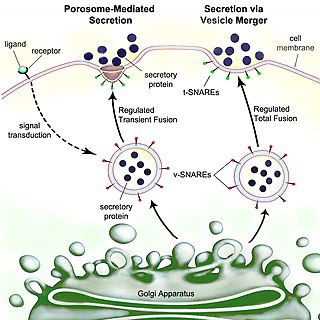
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.

Yersinia pseudotuberculosis is a Gram-negative bacterium that causes Far East scarlet-like fever in humans, who occasionally get infected zoonotically, most often through the food-borne route. Animals are also infected by Y. pseudotuberculosis. The bacterium is urease positive.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:

Intimin is a virulence factor (adhesin) of EPEC and EHEC E. coli strains. It is an attaching and effacing (A/E) protein, which with other virulence factors is necessary and responsible for enteropathogenic and enterohaemorrhagic diarrhoea.
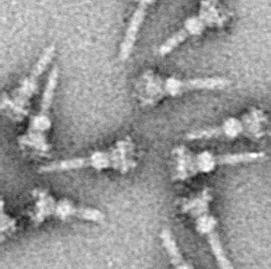
Type three secretion system is a protein appendage found in several Gram-negative bacteria.
Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

In molecular biology, LcrV is a protein found in Yersinia pestis and several other bacterial species. It forms part of the Yersinia pestis virulence protein factors that also includes all Yops, or Yersinia outer protein, but the name has been kept out of convention. LcrV's main function is not actually known, but it is essential for the production of other Yops.

In molecular biology, YopH, N-terminal refers to an evolutionary conserved protein domain. This entry represents the N-terminal domain of YopH protein tyrosine phosphatase (PTP).

In molecular biology, trimeric autotransporter adhesins (TAAs), are proteins found on the outer membrane of Gram-negative bacteria. Bacteria use TAAs in order to infect their host cells via a process called cell adhesion. TAAs also go by another name, oligomeric coiled-coil adhesins, which is shortened to OCAs. In essence, they are virulence factors, factors that make the bacteria harmful and infective to the host organism.

In molecular biology, the protein domain YopE refers to the secretion of virulence factors in Gram-negative bacteria involves transportation of the protein across two membranes to reach the cell exterior. It not only infects the host cell but also protects the bacteria. It undergoes several mechanisms to evade the host's immune system. This particular protein domain can be referred to as a Rho GTPase-activating protein (GAP).

In molecular biology, the protein domain TyeA is short for Translocation of Yops into eukaryotic cells A. It controls the release of Yersinia outer proteins (Yops) which help Yersinia evade the immune system. More specifically, it interacts with the bacterial protein YopN via hydrophobic residues located on the helices.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.
Bacterial effectors are proteins secreted by pathogenic bacteria into the cells of their host, usually using a type 3 secretion system (TTSS/T3SS), a type 4 secretion system (TFSS/T4SS) or a Type VI secretion system (T6SS). Some bacteria inject only a few effectors into their host’s cells while others may inject dozens or even hundreds. Effector proteins may have many different activities, but usually help the pathogen to invade host tissue, suppress its immune system, or otherwise help the pathogen to survive. Effector proteins are usually critical for virulence. For instance, in the causative agent of plague, the loss of the T3SS is sufficient to render the bacteria completely avirulent, even when they are directly introduced into the bloodstream. Gram negative microbes are also suspected to deploy bacterial outer membrane vesicles to translocate effector proteins and virulence factors via a membrane vesicle trafficking secretory pathway, in order to modify their environment or attack/invade target cells, for example, at the host-pathogen interface.
The type VI secretion system (T6SS) is molecular machine used by a wide range of Gram-negative bacterial species to transport effectors from the interior of a bacterial cell across the cellular envelope into an adjacent target cell. While often reported that the T6SS was discovered in 2006 by researchers studying the causative agent of cholera, Vibrio cholerae, the first study demonstrating that T6SS genes encode a protein export apparatus was actually published in 2004, in a study of protein secretion by the fish pathogen Edwardsiella tarda.
The type 2 secretion system is a type of protein secretion machinery found in various species of Gram-negative bacteria, including many human pathogens such as Pseudomonas aeruginosa and Vibrio cholerae. The type II secretion system is one of six protein secretory systems commonly found in Gram-negative bacteria, along with the type I, type III, and type IV secretion systems, as well as the chaperone/usher pathway, the autotransporter pathway/type V secretion system, and the type VI secretion system. Like these other systems, the type II secretion system enables the transport of cytoplasmic proteins across the lipid bilayers that make up the cell membranes of Gram-negative bacteria. Secretion of proteins and effector molecules out of the cell plays a critical role in signaling other cells and in the invasion and parasitism of host cells.

RNA thermometers regulate gene expression in response to temperature allowing pathogens like Yersinia to switch on silent genes after entering the host organism. Usually, RNA thermometers are located in the 5'UTR, but an intergenic RNA thermometer was found in Yersinia pseudotuberculosis. The LcrFRNA thermometer together with the thermo-labile YmoA protein activates synthesis of the most crucial virulence activator LcrF (VirF). The RNA thermosensor sequence is 100% identical in all human pathogenic Yersinia species.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.

Nucleomodulins are a family of bacterial proteins that enter the nucleus of eukaryotic cells.
















