Organohalide respiration (OHR) (previously named halorespiration or dehalorespiration) is the use of halogenated compounds as terminal electron acceptors in anaerobic respiration. Organohalide respiration can play a part in microbial biodegradation. The most common substrates are chlorinated aliphatics (PCE, TCE), chlorinated phenols and chloroform. Organohalide-respiring bacteria are highly diverse. This trait is found in some Campylobacterota, Thermodesulfobacteriota, Chloroflexota (green nonsulfur bacteria), low G+C gram positive Clostridia, and ultramicrobacteria.

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.

Catechol 1,2- dioxygenase is an enzyme that catalyzes the oxidative ring cleavage of catechol to form cis,cis-muconic acid:
The crotonase family comprises mechanistically diverse proteins that share a conserved trimeric quaternary structure, the core of which consists of 4 turns of a (beta/beta/alpha)n superhelix.
The enzyme selenocysteine lyase (SCL) (EC 4.4.1.16) catalyzes the chemical reaction
In enzymology, a 4-chlorobenzoate dehalogenase (EC 3.8.1.6) is an enzyme that catalyzes the chemical reaction
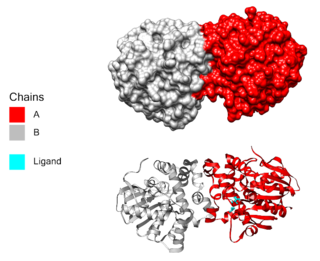
In enzymology, a haloacetate dehalogenase (EC 3.8.1.3) is an enzyme that catalyzes the chemical reaction
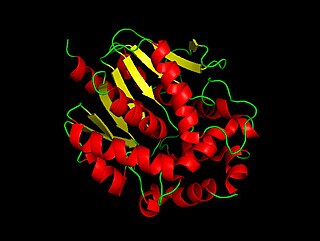
In enzymology, a haloalkane dehalogenase (EC 3.8.1.5) is an enzyme that catalyzes the chemical reaction
In enzymology, a (R)-2-haloacid dehalogenase(EC 3.8.1.9), DL-2-haloacid halidohydrolase (inversion of configuration), DL-DEXi, (R,S)-2-haloacid dehalogenase (configuration-inverting)) is an enzyme that catalyzes the chemical reaction
In enzymology, a (S)-2-haloacid dehalogenase (EC 3.8.1.2) is an enzyme that catalyzes the chemical reaction
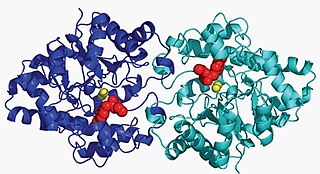
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:

Iodotyrosine deiodinase, also known as iodotyrosine dehalogenase 1, is a type of deiodinase enzyme that scavenges iodide by removing it from iodinated tyrosine residues in the thyroid gland. These iodinated tyrosines are produced during thyroid hormone biosynthesis. The iodide that is scavenged by iodotyrosine deiodinase is necessary to again synthesize the thyroid hormones. After synthesis, the thyroid hormones circulate through the body to regulate metabolic rate, protein expression, and body temperature. Iodotyrosine deiodinase is thus necessary to keep levels of both iodide and thyroid hormones in balance.
Dehalobacter is a genus in the phylum Bacillota (Bacteria).

The haloacid dehydrogenase superfamily is a superfamily of enzymes that include phosphatases, phosphonatases, P-type ATPases, beta-phosphoglucomutases, phosphomannomutases, and dehalogenases, and are involved in a variety of cellular processes ranging from amino acid biosynthesis to detoxification.
2-haloacid dehalogenase (configuration-retaining) (EC 3.8.1.11, 2-haloalkanoic acid dehalogenase, 2-haloalkanoid acid halidohydrolase, DL-2-haloacid dehalogenase, DL-DEXr) is an enzyme with systematic name (S)-2-haloacid dehalogenase (configuration-retaining). This enzyme catalyses the following chemical reaction
Achromobacter obae is a bacterium from the genus Achromobacter which contains the enzyme alpha-amino-epsilon-caprolactam racemase. The complete genome of A. obae has been sequenced.
Ancylobacter aquaticus is a bacterium from the family of Xanthobacteraceae which has been isolated from lake water in Copenhagen in Denmark. Ancylobacter aquaticus can degrade 1,2-dichloroethane and produces haloalkane dehalogenase.
Adsorbable Organic Halides (AOX) is a measure of the organic halogen load at a sampling site such as soil from a land fill, water, or sewage waste. The procedure measures chlorine, bromine, and iodine as equivalent halogens, but does not measure fluorine levels in the sample.
Reductive dehaholagenses (EC 1.97.1.8) are a group of enzymes utilized in organohalide respiring bacteria. These enzymes are mostly attached to the periplasmic side of the cytoplasmic membrane and play a central role in energy-conserving respiratory process for organohalide respiring bacteria by reducing organohalides. During such reductive dehalogenation reaction, organohalides are used as terminal electron acceptors. They catalyze the following general reactions:
Sam Hay is a chemist from New Zealand and a Reader in the Department of Chemistry at The University of Manchester. His research in general is based on computational chemistry and theoretical chemistry, specifically on the areas of In silico Enzymology, quantum mechanics roles in biological processes, kinetic modelling of complex reactions and high pressure spectroscopy.







