
Heterocysts or heterocytes are specialized nitrogen-fixing cells formed during nitrogen starvation by some filamentous cyanobacteria, such as Nostoc punctiforme, Cylindrospermum stagnale, and Anabaena sphaerica. They fix nitrogen from dinitrogen (N2) in the air using the enzyme nitrogenase, in order to provide the cells in the filament with nitrogen for biosynthesis.

Azotobacter is a genus of usually motile, oval or spherical bacteria that form thick-walled cysts and may produce large quantities of capsular slime. They are aerobic, free-living soil microbes that play an important role in the nitrogen cycle in nature, binding atmospheric nitrogen, which is inaccessible to plants, and releasing it in the form of ammonium ions into the soil. In addition to being a model organism for studying diazotrophs, it is used by humans for the production of biofertilizers, food additives, and some biopolymers. The first representative of the genus, Azotobacter chroococcum, was discovered and described in 1901 by Dutch microbiologist and botanist Martinus Beijerinck. Azotobacter species are Gram-negative bacteria found in neutral and alkaline soils, in water, and in association with some plants.
Paenarthrobacter ureafaciens KI72, popularly known as nylon-eating bacteria, is a strain of Paenarthrobacter ureafaciens that can digest certain by-products of nylon 6 manufacture. It uses a set of enzymes to digest nylon, popularly known as nylonase.
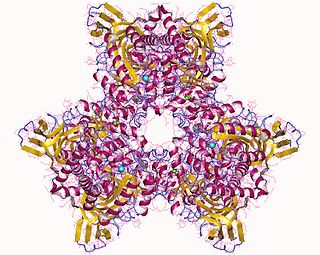
Atrazine Chlorohydrolase (AtzA) is an enzyme (E.C.3.8.1.8), which catalyzes the conversion of atrazine to hydroxyatrazine. Bacterial degradation determines the environmental impact and efficacy of an herbicide or pesticide. Initially, most pesticides are highly effective and show minimal bacterial degradation; however, bacteria can rapidly evolve and gain the ability to metabolize potential nutrients in the environment. Despite a remarkable structural similarity, degradation of atrazine by bacteria capable of melamine degradation was rare; however, since its introduction as a pesticide in the United States, bacteria capable of atrazine degradation have evolved. Currently, Pseudomonas sp. strain ADP seems to be the optimal bacterial strain for atrazine degradations, which appears to be the sole nitrogen source for the bacteria.

Glutamate carboxypeptidase is an enzyme. This enzyme catalyses the following chemical reaction
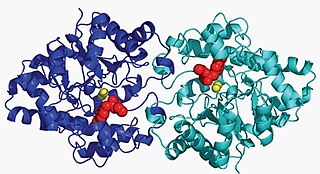
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:
Butyrivibrio is a genus of bacteria in Class Clostridia. Bacteria of this genus are common in the gastrointestinal systems of many animals. Genus Butyrivibrio was first described by Bryant and Small (1956) as anaerobic, butyric acid-producing, curved rods. Butyrivibrio cells are small, typically 0.4 – 0.6 µm by 2 – 5 µm. They are motile, using a single polar or subpolar monotrichous flagellum. They are commonly found singly or in short chains but it is not unusual for them to form long chains. Despite historically being described as Gram-negative, their cell walls contain derivatives of teichoic acid, and electron microscopy indicates that bacteria of this genus have a Gram-positive cell wall type. It is thought that they appear Gram-negative when Gram stained because their cell walls thin to 12 to 18 nm as they reach stationary phase.
2-Hydroxymuconate-6-semialdehyde dehydrogenase (EC 1.2.1.85, xylG [gene], praB [gene] ) is an enzyme with systematic name (2E,4Z)-2-hydroxy-6-oxohexa-2,4-dienoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
5-nitrosalicylate dioxygenase (EC 1.13.11.64, naaB (gene)) is an enzyme with systematic name 5-nitrosalicylate:oxygen 1,2-oxidoreductase (decyclizing). This enzyme catalyses the following chemical reaction
4-nitrocatechol 4-monooxygenase (EC 1.14.13.166) is an enzyme with systematic name 4-nitrocatechol,NAD(P)H:oxygen 4-oxidoreductase (4-hydroxylating, nitrite-forming). This enzyme catalyses the following chemical reaction:
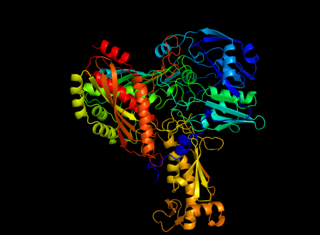
Caffeine dehydrogenase, commonly referred to in scientific literature as caffeine oxidase, is an enzyme with the systematic name caffeine:ubiquinone oxidoreductase. The enzyme is most well known for its ability to directly oxidize caffeine, a type of methylxanthine, to trimethyluric acid. Caffeine dehydrogenase can be found in bacterium Pseudomonas sp. CBB1 and in several species within the genera Alcaligenes, Rhodococcus, and Klebsiella.
The enzyme pyrethroid hydrolase (EC 3.1.1.88, pyrethroid-hydrolyzing carboxylesterase, pyrethroid-hydrolysing esterase, pyrethroid-hydrolyzing esterase, pyrethroid-selective esterase, pyrethroid-cleaving enzyme, permethrinase, PytH, EstP; systematic name pyrethroid-ester hydrolase) catalyses the reaction
The enzyme 2-oxo-3-(5-oxofuran-2-ylidene)propanoate lactonase (EC 3.1.1.91, naaC (gene); systematic name 2-oxo-3-(5-oxofuran-2-ylidene)propanoate lactonohydrolase) catalyses the reaction
Oligopeptidase A is an enzyme. This enzyme catalyses the following chemical reaction
Thauera aromatica is a species of bacteria. Its type strain is K 172T.
Azoarcus evansii is a species of bacteria. Its type strain is KB 740T.
Alcanivorax pacificus is a pyrene-degrading marine gammaproteobacterium. It is of the genus Alcanivorax, a group of marine bacteria known for degrading hydrocarbons. When originally proposed, the genus Alcanivorax comprised six distinguishable species. However, A. pacificus, a seventh strain, was isolated from deep sea sediments in the West Pacific Ocean by Shanghai Majorbio Bio-pharm Technology Co., Ltd. in 2011. A. pacificus’s ability to degrade hydrocarbons can be employed for cleaning up oil-contaminated oceans through bioremediation. The genomic differences present in this strain of Alcanivorax that distinguish it from the original consortium are important to understand to better utilize this bacteria for bioremediation.
Sphingobium japonicum is a hexachlorocyclohexane-degrading bacteria with type strain MTCC 6362T. Its genome has been sequenced.

Cyanothece is a genus of unicellular, diazotrophic, oxygenic photosynthesizing cyanobacteria.

The species Rhizorhabdus wittichii, formerly Sphingomonas wittichii, is a Gram-negative, rod-shaped motile bacterium, with an optimum growth temperature at 30 °C. It forms a greyish white colony. It has been found to have a 67mol% of DNA G+C content.








