Related Research Articles

A pilus is a hair-like cell-surface appendage found on many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric.

Escherichia coli ( ESH-ə-RIK-ee-ə KOH-lye) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC and ETEC are pathogenic, can cause serious food poisoning in their hosts and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

Staphylococcus aureus is a gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe, meaning that it can grow without oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA). The bacterium is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

In biological classification, subspecies is a rank below species, used for populations that live in different areas and vary in size, shape, or other physical characteristics (morphology), but that can successfully interbreed. Not all species have subspecies, but for those that do there must be at least two. Subspecies is abbreviated as subsp. or ssp. and the singular and plural forms are the same.

Avian influenza, also known as avian flu or bird flu, is a disease caused by the influenza A virus, which primarily affects birds but can sometimes affect mammals including humans. Wild aquatic birds are the primary host of the influenza A virus, which is enzootic in many bird populations.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

The laboratory mouse or lab mouse is a small mammal of the order Rodentia which is bred and used for scientific research or feeders for certain pets. Laboratory mice are usually of the species Mus musculus. They are the most commonly used mammalian research model and are used for research in genetics, physiology, psychology, medicine and other scientific disciplines. Mice belong to the Euarchontoglires clade, which includes humans. This close relationship, the associated high homology with humans, their ease of maintenance and handling, and their high reproduction rate, make mice particularly suitable models for human-oriented research. The laboratory mouse genome has been sequenced and many mouse genes have human homologues. Lab mice are sold at pet stores for snake food and can also be kept as pets.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.
Heterothallic species have sexes that reside in different individuals. The term is applied particularly to distinguish heterothallic fungi, which require two compatible partners to produce sexual spores, from homothallic ones, which are capable of sexual reproduction from a single organism.
Complementation refers to a genetic process when two strains of an organism with different homozygous recessive mutations that produce the same mutant phenotype have offspring that express the wild-type phenotype when mated or crossed. Complementation will ordinarily occur if the mutations are in different genes. Complementation may also occur if the two mutations are at different sites within the same gene, but this effect is usually weaker than that of intergenic complementation. When the mutations are in different genes, each strain's genome supplies the wild-type allele to "complement" the mutated allele of the other strain's genome. Since the mutations are recessive, the offspring will display the wild-type phenotype. A complementation test can test whether the mutations in two strains are in different genes. Complementation is usually weaker or absent if the mutations are in the same gene. The convenience and essence of this test is that the mutations that produce a phenotype can be assigned to different genes without the exact knowledge of what the gene product is doing on a molecular level. American geneticist Edward B. Lewis developed the complementation test.
The International Code of Nomenclature of Prokaryotes (ICNP) or Prokaryotic Code, formerly the International Code of Nomenclature of Bacteria (ICNB) or Bacteriological Code (BC), governs the scientific names for Bacteria and Archaea. It denotes the rules for naming taxa of bacteria, according to their relative rank. As such it is one of the nomenclature codes of biology.

Shigella boydii is a Gram-negative bacterium of the genus Shigella. Like other members of the genus, S. boydii is a nonmotile, nonsporeforming, rod-shaped bacterium which can cause dysentery in humans through fecal-oral contamination.

Fungi are a diverse group of organisms that employ a huge variety of reproductive strategies, ranging from fully asexual to almost exclusively sexual species. Most species can reproduce both sexually and asexually, alternating between haploid and diploid forms. This contrasts with most multicellular eukaryotes such as mammals, where the adults are usually diploid and produce haploid gametes which combine to form the next generation. In fungi, both haploid and diploid forms can reproduce – haploid individuals can undergo asexual reproduction while diploid forms can produce gametes that combine to give rise to the next generation.
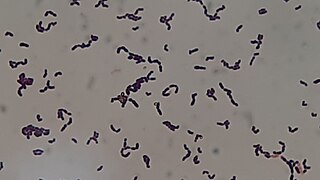
Streptococcus sanguinis, formerly known as Streptococcus sanguis, is a Gram-positive facultative anaerobic coccus species of bacteria and a member of the Viridans Streptococcus group. S. sanguinis is a normal inhabitant of the healthy human mouth where it is particularly found in dental plaque, where it modifies the environment to make it less hospitable for other strains of Streptococcus that cause cavities, such as Streptococcus mutans.

The genetic structure of H5N1, a highly pathogenic avian influenza virus, is characterized by a segmented RNA genome consisting of eight gene segments that encode for various viral proteins essential for replication, host adaptation, and immune evasion.
Capnocytophaga is a genus of Gram-negative bacteria. Normally found in the oropharyngeal tract of mammals and are involved in the pathogenesis of some animal bite wounds and periodontal diseases.

Cannabis strains is a popular name to refer to plant varieties of the monospecific genus Cannabis sativa L.. They are either pure or hybrid varieties of the plant, which encompasses various sub-species C. sativa, C. indica, and C. ruderalis.

Fission, in biology, is the division of a single entity into two or more parts and the regeneration of those parts to separate entities resembling the original. The object experiencing fission is usually a cell, but the term may also refer to how organisms, bodies, populations, or species split into discrete parts. The fission may be binary fission, in which a single organism produces two parts, or multiple fission, in which a single entity produces multiple parts.
SCCmec, or staphylococcal cassette chromosome mec, is a mobile genetic element of Staphylococcus bacterial species. This genetic sequence includes the mecA gene coding for resistance to the antibiotic methicillin and is the only known way for Staphylococcus strains to spread the gene in the wild by horizontal gene transfer. SCCmec is a 21 to 60 kb long genetic element that confers broad-spectrum β-lactam resistance to MRSA. Moreover, additional genetic elements like Tn554, pT181, and pUB110 can be found in SCCmec, which have the capability to render resistance to various non-β-lactam drugs.
This glossary of virology is a list of definitions of terms and concepts used in virology, the study of viruses, particularly in the description of viruses and their actions. Related fields include microbiology, molecular biology, and genetics.
References
- ↑ Prat L, Heinemann IU, Aerni HR, Rinehart J, O'Donoghue P, Söll D (Dec 2012). "Carbon source-dependent expansion of the genetic code in bacteria". Proceedings of the National Academy of Sciences of the United States of America. 109 (51): 21070–5. Bibcode:2012PNAS..10921070P. doi: 10.1073/pnas.1218613110 . PMC 3529041 . PMID 23185002.