
A pseudopod or pseudopodium is a temporary arm-like projection of a eukaryotic cell membrane that is emerged in the direction of movement. Filled with cytoplasm, pseudopodia primarily consist of actin filaments and may also contain microtubules and intermediate filaments. Pseudopods are used for motility and ingestion. They are often found in amoebas.

Microfilaments, also called actin filaments, are protein filaments in the cytoplasm of eukaryotic cells that form part of the cytoskeleton. They are primarily composed of polymers of actin, but are modified by and interact with numerous other proteins in the cell. Microfilaments are usually about 7 nm in diameter and made up of two strands of actin. Microfilament functions include cytokinesis, amoeboid movement, cell motility, changes in cell shape, endocytosis and exocytosis, cell contractility, and mechanical stability. Microfilaments are flexible and relatively strong, resisting buckling by multi-piconewton compressive forces and filament fracture by nanonewton tensile forces. In inducing cell motility, one end of the actin filament elongates while the other end contracts, presumably by myosin II molecular motors. Additionally, they function as part of actomyosin-driven contractile molecular motors, wherein the thin filaments serve as tensile platforms for myosin's ATP-dependent pulling action in muscle contraction and pseudopod advancement. Microfilaments have a tough, flexible framework which helps the cell in movement.

The Wiskott–Aldrich syndrome protein (WASp) is a 502-amino acid protein expressed in cells of the hematopoietic system that in humans is encoded by the WAS gene. In the inactive state, WASp exists in an autoinhibited conformation with sequences near its C-terminus binding to a region near its N-terminus. Its activation is dependent upon CDC42 and PIP2 acting to disrupt this interaction, causing the WASp protein to 'open'. This exposes a domain near the WASp C-terminus that binds to and activates the Arp2/3 complex. Activated Arp2/3 nucleates new F-actin.

Podosomes are conical, actin-rich structures found on the outer surface of the plasma membrane of animal cells. Their size ranges from approximately 0.5 μm to 2.0 μm in diameter. While usually situated on the periphery of the cellular membrane, these unique structures display a polarized pattern of distribution in migrating cells, situating at the front border between the lamellipodium and lamellum. Their primary purpose is connected to cellular motility and invasion; therefore, they serve as both sites of attachment and degradation along the extracellular matrix. Many different specialized cells exhibit these dynamic structures such as invasive cancer cells, osteoclasts, vascular smooth muscle cells, endothelial cells, and certain immune cells like macrophages and dendritic cells.
The lamellipodium is a cytoskeletal protein actin projection on the leading edge of the cell. It contains a quasi-two-dimensional actin mesh; the whole structure propels the cell across a substrate. Within the lamellipodia are ribs of actin called microspikes, which, when they spread beyond the lamellipodium frontier, are called filopodia. The lamellipodium is born of actin nucleation in the plasma membrane of the cell and is the primary area of actin incorporation or microfilament formation of the cell.
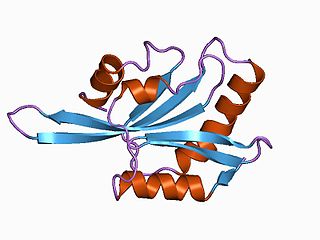
ADF/cofilin is a family of actin-binding proteins associated with the rapid depolymerization of actin microfilaments that give actin its characteristic dynamic instability. This dynamic instability is central to actin's role in muscle contraction, cell motility and transcription regulation.
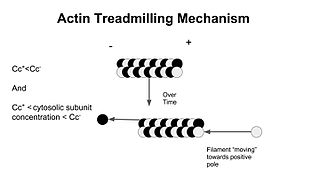
In molecular biology, treadmilling is a phenomenon observed within protein filaments of the cytoskeletons of many cells, especially in actin filaments and microtubules. It occurs when one end of a filament grows in length while the other end shrinks, resulting in a section of filament seemingly "moving" across a stratum or the cytosol. This is due to the constant removal of the protein subunits from these filaments at one end of the filament, while protein subunits are constantly added at the other end. Treadmilling was discovered by Wegner, who defined the thermodynamic and kinetic constraints. Wegner recognized that: “The equilibrium constant (K) for association of a monomer with a polymer is the same at both ends, since the addition of a monomer to each end leads to the same polymer.”; a simple reversible polymer can’t treadmill; ATP hydrolysis is required. GTP is hydrolyzed for microtubule treadmilling.

Cortactin is a monomeric protein located in the cytoplasm of cells that can be activated by external stimuli to promote polymerization and rearrangement of the actin cytoskeleton, especially the actin cortex around the cellular periphery. It is present in all cell types. When activated, it will recruit Arp2/3 complex proteins to existing actin microfilaments, facilitating and stabilizing nucleation sites for actin branching. Cortactin is important in promoting lamellipodia formation, invadopodia formation, cell migration, and endocytosis.
In cell biology, microtubule nucleation is the event that initiates de novo formation of microtubules (MTs). These filaments of the cytoskeleton typically form through polymerization of α- and β-tubulin dimers, the basic building blocks of the microtubule, which initially interact to nucleate a seed from which the filament elongates.
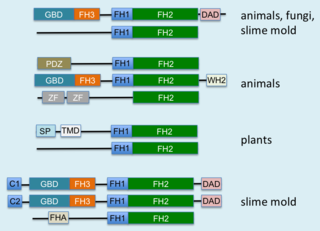
Formins (formin homology proteins) are a group of proteins that are involved in the polymerization of actin and associate with the fast-growing end (barbed end) of actin filaments. Most formins are Rho-GTPase effector proteins. Formins regulate the actin and microtubule cytoskeleton and are involved in various cellular functions such as cell polarity, cytokinesis, cell migration and SRF transcriptional activity. Formins are multidomain proteins that interact with diverse signalling molecules and cytoskeletal proteins, although some formins have been assigned functions within the nucleus.

Stress fibers are contractile actin bundles found in non-muscle cells. They are composed of actin (microfilaments) and non-muscle myosin II (NMMII), and also contain various crosslinking proteins, such as α-actinin, to form a highly regulated actomyosin structure within non-muscle cells. Stress fibers have been shown to play an important role in cellular contractility, providing force for a number of functions such as cell adhesion, migration and morphogenesis.

Invadopodia are actin-rich protrusions of the plasma membrane that are associated with degradation of the extracellular matrix in cancer invasiveness and metastasis. Very similar to podosomes, invadopodia are found in invasive cancer cells and are important for their ability to invade through the extracellular matrix, especially in cancer cell extravasation. Invadopodia are generally visualized by the holes they create in ECM -coated plates, in combination with immunohistochemistry for the invadopodia localizing proteins such as cortactin, actin, Tks5 etc. Invadopodia can also be used as a marker to quantify the invasiveness of cancer cell lines in vitro using a hyaluronic acid hydrogel assay.

Protein cordon-bleu is a protein that in humans is encoded by the COBL gene.

The Actin assembly-inducing protein (ActA) is a protein encoded and used by Listeria monocytogenes to propel itself through a mammalian host cell. ActA is a bacterial surface protein comprising a membrane-spanning region. In a mammalian cell the bacterial ActA interacts with the Arp2/3 complex and actin monomers to induce actin polymerization on the bacterial surface generating an actin comet tail. The gene encoding ActA is named actA or prtB.
Actin remodeling is the biochemical process that allows for the dynamic alterations of cellular organization. The remodeling of actin filaments occurs in a cyclic pattern on cell surfaces and exists as a fundamental aspect to cellular life. During the remodeling process, actin monomers polymerize in response to signaling cascades that stem from environmental cues. The cell's signaling pathways cause actin to affect intracellular organization of the cytoskeleton and often consequently, the cell membrane. Again triggered by environmental conditions, actin filaments break back down into monomers and the cycle is completed. Actin-binding proteins (ABPs) aid in the transformation of actin filaments throughout the actin remodeling process. These proteins account for the diverse structure and changes in shape of Eukaryotic cells. Despite its complexity, actin remodeling may result in complete cytoskeletal reorganization in under a minute.
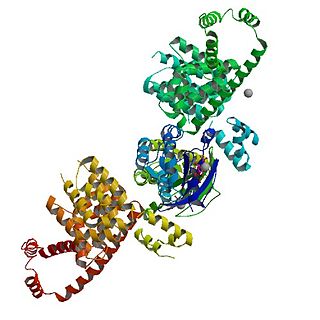
mDia1 is a member of the protein family called the formins and is a Rho effector. It is the mouse version of the diaphanous homolog 1 of Drosophila. mDia1 localizes to cells' mitotic spindle and midbody, plays a role in stress fiber and filopodia formation, phagocytosis, activation of serum response factor, formation of adherens junctions, and it can act as a transcription factor. mDia1 accelerates actin nucleation and elongation by interacting with barbed ends of actin filaments. The gene encoding mDia1 is located on Chromosome 18 of Mus musculus and named Diap1.

Formin-like protein 3 (FMNL3), also known as WW domain-binding protein 3 (WBP-3), is a protein that in humans is encoded by the FMNL3 gene.

Arp2/3 complex is a seven-subunit protein complex that plays a major role in the regulation of the actin cytoskeleton. It is a major component of the actin cytoskeleton and is found in most actin cytoskeleton-containing eukaryotic cells. Two of its subunits, the Actin-Related Proteins ARP2 and ARP3, closely resemble the structure of monomeric actin and serve as nucleation sites for new actin filaments. The complex binds to the sides of existing ("mother") filaments and initiates growth of a new ("daughter") filament at a distinctive 70-degree angle from the mother. Branched actin networks are created as a result of this nucleation of new filaments. The regulation of rearrangements of the actin cytoskeleton is important for processes like cell locomotion, phagocytosis, and intracellular motility of lipid vesicles.
Paracytophagy is the cellular process whereby a cell engulfs a protrusion which extends from a neighboring cell. This protrusion may contain material which is actively transferred between the cells. The process of paracytophagy was first described as a crucial step during cell-to-cell spread of the intracellular bacterial pathogen Listeria monocytogenes, and is also commonly observed in Shigella flexneri. Paracytophagy allows these intracellular pathogens to spread directly from cell to cell, thus escaping immune detection and destruction. Studies of this process have contributed significantly to our understanding of the role of the actin cytoskeleton in eukaryotic cells.
SPIRE1 is a protein that interacts with actin monomers and actin nucleating formin proteins. SPIRE1 was first identified in Drosophila melanogaster. SPIRE1 contains an N-terminal KIND domain which binds formins and four actin-binding WH2 domains which nucleate actin filaments.














