Bioinformatics is an interdisciplinary field that develops methods and software tools for understanding biological data. As an interdisciplinary field of science, bioinformatics combines biology, computer science, information engineering, mathematics and statistics to analyze and interpret biological data. Bioinformatics has been used for in silico analyses of biological queries using mathematical and statistical techniques.

A barcode is an optical, machine-readable representation of data; the data usually describes something about the object that carries the barcode. Traditional barcodes systematically represent data by varying the widths and spacings of parallel lines, and may be referred to as linear or one-dimensional (1D). Later, two-dimensional (2D) variants were developed, using rectangles, dots, hexagons and other geometric patterns, called matrix codes or 2D barcodes, although they do not use bars as such. Initially, barcodes were only scanned by special optical scanners called barcode readers. Later application software became available for devices that could read images, such as smartphones with cameras.

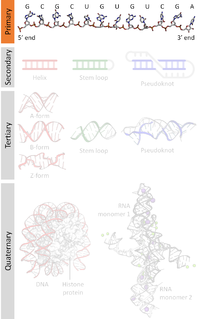
A DNA sequencer is a scientific instrument used to automate the DNA sequencing process. Given a sample of DNA, a DNA sequencer is used to determine the order of the four bases: G (guanine), C (cytosine), A (adenine) and T (thymine). This is then reported as a text string, called a read. Some DNA sequencers can be also considered optical instruments as they analyze light signals originating from fluorochromes attached to nucleotides.

The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). The NCBI is located in Bethesda, Maryland and was founded in 1988 through legislation sponsored by Senator Claude Pepper.
Bold is a font style used for emphasis.

DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.
Internal transcribed spacer (ITS) refers to the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript.

Astraptes fulgerator, the two-barred flasher, is a cryptic species complex in the spread-wing skipper butterfly genus Astraptes. It ranges all over the Americas, from the southern United States to northern Argentina.
The Consortium for the Barcode of Life (CBOL) is an international initiative dedicated to supporting the development of DNA barcoding as a global standard for species identification. CBOL's Secretariat Office is hosted by the National Museum of Natural History, Smithsonian Institution, in Washington, DC. Barcoding was proposed in 2003 by Prof. Paul Hebert of the University of Guelph in Ontario as a way of distinguishing and identifying species with a short standardized gene sequence. Hebert proposed the 648 bases of the Folmer region of the mitochondrial gene cytochrome-C oxidase-1 as the standard barcode region. Dr. Hebert is the Director of the Biodiversity Institute of Ontario, the Canadian Centre for DNA Barcoding, and the International Barcode of Life Project (iBOL), all headquartered at the University of Guelph. The Barcode of Life Data Systems (BOLD) is also located at the University of Guelph.

In biology, a species complex is a group of closely related organisms that are very similar in appearance to the point that the boundaries between them are often unclear. Terms sometimes used synonymously but with more precise meanings are: cryptic species for two or more species hidden under one species name, sibling species for two cryptic species that are each other's closest relative, and species flock for a group of closely related species living in the same habitat. As informal taxonomic ranks, species group, species aggregate, and superspecies are also in use.
454 Life Sciences was a biotechnology company based in Branford, Connecticut that specialized in high-throughput DNA sequencing. It was acquired by Roche in 2007 and shut down by Roche in 2013 when its technology became noncompetitive, although production continued until mid-2016.

The Intelligent Mail Barcode is a 65-bar barcode for use on mail in the United States. The term "Intelligent Mail" refers to services offered by the United States Postal Service for domestic mail delivery. The IM barcode is intended to provide greater information and functionality than its predecessors POSTNET and PLANET. An Intelligent Mail barcode has also been referred to as a One Code Solution and a 4-State Customer Barcode, abbreviated 4CB, 4-CB or USPS4CB. The complete specification can be found in USPS Document USPS-B-3200. It effectively incorporates the routing ZIP code and tracking information included in previously used postal barcode standards.
In biology, a species ( ) is the basic unit of classification and a taxonomic rank of an organism, as well as a unit of biodiversity. A species is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. Other ways of defining species include their karyotype, DNA sequence, morphology, behaviour or ecological niche. In addition, paleontologists use the concept of the chronospecies since fossil reproduction cannot be examined. While these definitions may seem adequate, when looked at more closely they represent problematic species concepts. For example, the boundaries between closely related species become unclear with hybridisation, in a species complex of hundreds of similar microspecies, and in a ring species. Also, among organisms that reproduce only asexually, the concept of a reproductive species breaks down, and each clone is potentially a microspecies.
Mitochondrial DNA is an academic journal that publishes review articles on the current and developing technologies around mitochondrial DNA research and discovery. It is published by Informa Healthcare.
Optical mapping is a technique for constructing ordered, genome-wide, high-resolution restriction maps from single, stained molecules of DNA, called "optical maps". By mapping the location of restriction enzyme sites along the unknown DNA of an organism, the spectrum of resulting DNA fragments collectively serves as a unique "fingerprint" or "barcode" for that sequence. Originally developed by Dr. David C. Schwartz and his lab at NYU in the 1990s this method has since been integral to the assembly process of many large-scale sequencing projects for both microbial and eukaryotic genomes. Later technologies use DNA melting, DNA competitive binding or enzymatic labelling in order to create the optical mappings.
Applied Food Technologies, Inc. (AFT) is a privately held corporation in Alachua, Florida that develops diagnostics needed in the seafood industry and runs fee-for-service species identification and verification programs. The DNA-based species identification diagnostics developed by AFT are used in-house and are also packaged in a kit format and sold to federal, state and private laboratories in the US, Europe and Asia. AFT has established an extensive library of taxonomically verified fish species and has developed DNA standards for these fish.

The Braconinae are a large subfamily of braconid parasitoid wasps with more than 2,000 described species. Many species, including Bracon brevicornis, have been used in biocontrol programs.
The Cardiochilinae are a subfamily of braconid parasitoid wasps. This subfamily has been treated as a tribe of Microgastrinae in the past. Some species including Toxoneuron nigriceps have been used in biocontrol programs.

Hachimoji DNA is a synthetic nucleic acid analog that uses four synthetic nucleotides in addition to the four present in the natural nucleic acids, DNA and RNA. This leads to four allowed base pairs: two unnatural base pairs formed by the synthetic nucleobases in addition to the two normal pairs. Hachimoji bases have been demonstrated in both DNA and RNA analogs, using deoxyribose and ribose respectively as the backbone sugar.