The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage.
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It is also a product and services market, with an estimated value of $168 billion by 2017.
Semisynthesis or partial chemical synthesis is a type of chemical synthesis that uses chemical compounds isolated from natural sources as the starting materials to produce other novel compounds with distinct chemical and medicinal properties. These novel compounds are generally high molecular weight or have complex molecular structure, more so than those produced by total synthesis from simple starting materials. Semisynthesis is a means of preparing many medicines more cheaply than if by total synthesis, because fewer chemical steps are necessary.
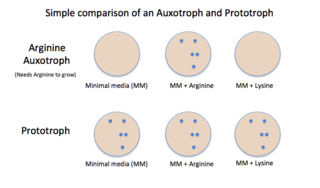
Auxotrophy is the inability of an organism to synthesize a particular organic compound required for its growth. An auxotroph is an organism that displays this characteristic; auxotrophic is the corresponding adjective. Auxotrophy is the opposite of prototrophy, which is characterized by the ability to synthesize all the compounds needed for growth.

A peptidomimetic is a small protein-like chain designed to mimic a peptide. They typically arise either from modification of an existing peptide, or by designing similar systems that mimic peptides, such as peptoids and β-peptides. Irrespective of the approach, the altered chemical structure is designed to advantageously adjust the molecular properties such as, stability or biological activity. This can have a role in the development of drug-like compounds from existing peptides. These modifications involve changes to the peptide that will not occur naturally. Based on their similarity with the precursor peptide, peptidomimetics can be grouped into four classes where A features the most and D the least similarities. Classes A and B involve peptide-like scaffolds, while classes C and D include small molecules.
Metabolic engineering is the practice of optimizing genetic and regulatory processes within cells to increase the cells' production of a certain substance. These processes are chemical networks that use a series of biochemical reactions and enzymes that allow cells to convert raw materials into molecules necessary for the cell’s survival. Metabolic engineering specifically seeks to mathematically model these networks, calculate a yield of useful products, and pin point parts of the network that constrain the production of these products. Genetic engineering techniques can then be used to modify the network in order to relieve these constraints. Once again this modified network can be modeled to calculate the new product yield.
Chemical biology is a scientific discipline spanning the fields of chemistry and biology. The discipline involves the application of chemical techniques, analysis, and often small molecules produced through synthetic chemistry, to the study and manipulation of biological systems. In contrast to biochemistry, which involves the study of the chemistry of biomolecules and regulation of biochemical pathways within and between cells, chemical biology deals with chemistry applied to biology.
Threose nucleic acid (TNA) is an artificial genetic polymer invented by Albert Eschenmoser. TNA has a backbone structure composed of repeating threose sugars linked together by phosphodiester bonds. Like DNA and RNA, TNA can store genetic information in strings of nucleotide sequences. TNA is not known to occur naturally and is synthesized chemically in the laboratory under controlled conditions. It is believed by some that TNA could be an evolutionary pathway to RNA.

Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis, selection, and amplification. It can be performed in vivo(in living organisms), or in vitro. Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as studies of fundamental evolutionary principles in a controlled, laboratory environment.

In pharmacology, the term mechanism of action (MOA) refers to the specific biochemical interaction through which a drug substance produces its pharmacological effect. A mechanism of action usually includes mention of the specific molecular targets to which the drug binds, such as an enzyme or receptor. Receptor sites have specific affinities for drugs based on the chemical structure of the drug, as well as the specific action that occurs there. Drugs that do not bind to receptors produce their corresponding therapeutic effect by simply interacting with chemical or physical properties in the body. Common examples of drugs that work in this way are antacids and laxatives.

Fusion proteins or chimeric (\kī-ˈmir-ik) proteins are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this fusion gene results in a single or multiple polypeptides with functional properties derived from each of the original proteins. Recombinant fusion proteins are created artificially by recombinant DNA technology for use in biological research or therapeutics. Chimeric or chimera usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. Chimeric mutant proteins occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncoproteins. The bcr-abl fusion protein is a well-known example of an oncogenic fusion protein, and is considered to be the primary oncogenic driver of chronic myelogenous leukemia.

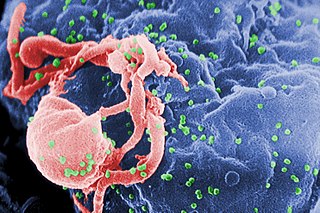
HIV-1 protease (PR) is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS. HIV protease cleaves newly synthesized polyproteins at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell. Without effective HIV protease, HIV virions remain uninfectious.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE) or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.

Frances Hamilton Arnold is an American chemical engineer and Nobel Laureate. She is the Linus Pauling Professor of Chemical Engineering, Bioengineering and Biochemistry at the California Institute of Technology (Caltech). In 2018, she was awarded the Nobel Prize in Chemistry for pioneering the use of directed evolution to engineer enzymes.
Glycorandomization, is a drug discovery and drug development technology platform to enable the rapid diversification of bioactive small molecules, drug leads and/or approved drugs through the attachment of sugars. Initially developed as a facile method to manipulate carbohydrate substitutions of naturally occurring glycosides to afford the corresponding differentially glycosylated natural product libraries, glycorandomization applications have expanded to include both small molecules and even macromolecules (proteins). Also referred to as 'glycodiversification', glycorandomization has led to the discovery of new glycoside analogs which display improvements in potency, selectivity and/or ADMET as compared to the parent molecule.
Chemical genetics is the investigation of the function of proteins and signal transduction pathways in cells by the screening of chemical libraries of small molecules. Chemical genetics is analogous to classical genetic screen where random mutations are introduced in organisms, the phenotype of these mutants is observed, and finally the specific gene mutation (genotype) that produced that phenotype is identified. In chemical genetics, the phenotype is disturbed not by introduction of mutations, but by exposure to small molecule tool compounds. Phenotypic screening of chemical libraries is used to identify drug targets or to validate those targets in experimental models of disease. Recent applications of this topic have been implicated in signal transduction, which may play a role in discovering new cancer treatments. Chemical genetics can serve as a unifying study between chemistry and biology. The approach was first proposed by Tim Mitchison in 1994 in an opinion piece in the journal Chemistry & Biology entitled "Towards a pharmacological genetics".
Synthetic immunology is the rational design and construction of synthetic systems that perform complex immunological functions. Functions include using specific cell markers to target cells for destruction and or interfering with immune reactions. US Food and Drug Administration (FDA)-approved immune system modulators include anti-inflammatory and immunosuppressive agents, vaccines, therapeutic antibodies and Toll-like receptor (TLR) agonists.

Epistasis is the phenomenon where the effect of one gene (locus) is dependent on the presence of one or more 'modifier genes', i.e. the genetic background. Originally the term meant that the phenotypic effect of one gene is masked by a different gene (locus). Thus, epistatic mutations have different effects in combination than individually. It was originally a concept from genetics but is now used in biochemistry, computational biology and evolutionary biology. It arises due to interactions, either between genes, or within them, leading to non-linear effects. Epistasis has a large influence on the shape of evolutionary landscapes, which leads to profound consequences for evolution and evolvability of phenotypic traits.

Strigolactones are a group of chemical compounds produced by a plant's roots. Due to their mechanism of action, these molecules have been classified as plant hormones or phytohormones. So far, strigolactones have been identified to be responsible for three different physiological processes: First, they promote the germination of parasitic organisms that grow in the host plant's roots, such as Strigalutea and other plants of the genus Striga. Second, strigolactones are fundamental for the recognition of the plant by symbiotic fungi, especially arbuscular mycorrhizal fungi, because they establish a mutualistic association with these plants, and provide phosphate and other soil nutrients. Third, strigolactones have been identified as branching inhibition hormones in plants; when present, these compounds prevent excess bud growing in stem terminals, stopping the branching mechanism in plants.
RNA-targeting small molecules represent a class of small molecules, organic compounds with traditional drug properties that can bind to RNA secondary or tertiary structures and alter translation patterns, localization, and degradation.