Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.
Bookmarking refers to a potential mechanism of transmission of gene expression programs through cell division.

GATA-binding factor 1 or GATA-1 is the founding member of the GATA family of transcription factors. This protein is widely expressed throughout vertebrate species. In humans and mice, it is encoded by the GATA1 and Gata1 genes, respectively. These genes are located on the X chromosome in both species.

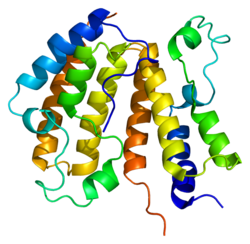
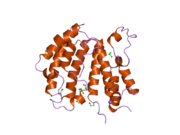
A bromodomain is an approximately 110 amino acid protein domain that recognizes acetylated lysine residues, such as those on the N-terminal tails of histones. Bromodomains, as the "readers" of lysine acetylation, are responsible in transducing the signal carried by acetylated lysine residues and translating it into various normal or abnormal phenotypes. Their affinity is higher for regions where multiple acetylation sites exist in proximity. This recognition is often a prerequisite for protein-histone association and chromatin remodeling. The domain itself adopts an all-α protein fold, a bundle of four alpha helices each separated by loop regions of variable lengths that form a hydrophobic pocket that recognizes the acetyl lysine.

Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation.

Histone H4 is a protein that in humans is encoded by the HIST4H4 gene.

Interferon regulatory factor 2 is a protein that in humans is encoded by the IRF2 gene.

Bromodomain-containing protein 2 is a protein that in humans is encoded by the BRD2 gene. BRD2 is part of the Bromodomain and Extra-Terminal motif (BET) protein family that also contains BRD3, BRD4, and BRDT in mammals

Metastasis-associated protein MTA2 is a protein that in humans is encoded by the MTA2 gene.

Homeobox protein Hox-B2 is a protein that in humans is encoded by the HOXB2 gene.

Transcription factor NF-E2 45 kDa subunit is a protein that in humans is encoded by the NFE2 gene.

Bromodomain-containing protein 4 is a protein that in humans is encoded by the BRD4 gene.

Bromodomain-containing protein 7 is a protein that in humans is encoded by the BRD7 gene.

Transcription factor MafK is a bZip Maf transcription factor protein that in humans is encoded by the MAFK gene.

ASH1L is a histone-lysine N-methyltransferase enzyme encoded by the ASH1L gene located at chromosomal band 1q22. ASH1L is the human homolog of Drosophila Ash1.

Bromodomain testis-specific protein is a protein that in humans is encoded by the BRDT gene. It is a member of the Bromodomain and Extra-terminal motif (BET) protein family.
Ming-Ming Zhou, Ph.D., is an internationally renowned expert in structural and chemical biology, NMR spectroscopy and drug design. He is currently the Dr. Harold and Golden Lamport Professor and Chairman of the Department of Pharmacological Sciences and Co-Director of the Drug Discovery Institute at the Icahn School of Medicine at Mount Sinai and Mount Sinai Health System in New York City as well as Professor of Oncological Sciences.

JQ1 is a thienotriazolodiazepine and a potent inhibitor of the BET family of bromodomain proteins which include BRD2, BRD3, BRD4, and the testis-specific protein BRDT in mammals. BET inhibitors structurally similar to JQ1 are being tested in clinical trials for a variety of cancers including NUT midline carcinoma. It was developed by the James Bradner laboratory at Brigham and Women's Hospital and named after chemist Jun Qi. The chemical structure was inspired by patent of similar BET inhibitors by Mitsubishi Tanabe Pharma [WO/2009/084693]. Structurally it is related to benzodiazepines. While widely used in laboratory applications, JQ1 is not itself being used in human clinical trials because it has a short half life.
BET inhibitors are a class of drugs that reversibly bind the bromodomains of Bromodomain and Extra-Terminal motif (BET) proteins BRD2, BRD3, BRD4, and BRDT, and prevent protein-protein interaction between BET proteins and acetylated histones and transcription factors.

In genetics, transcriptional amplification is the process in which the total amounts of messenger RNA (mRNA) molecules from expressed genes are increased during disease, development, or in response to stimuli.