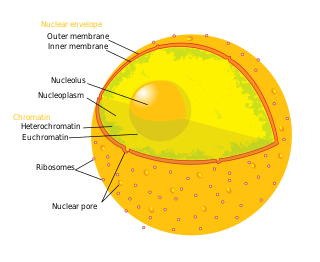
A nuclear pore is a part of a large complex of proteins, known as a nuclear pore complex that spans the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are approximately 1,000 nuclear pore complexes (NPCs) in the nuclear envelope of a vertebrate cell, but this number varies depending on cell type and the stage in the life cycle. The human nuclear pore complex (hNPC) is a 110 megadalton (MDa) structure. The proteins that make up the nuclear pore complex are known as nucleoporins; each NPC contains at least 456 individual protein molecules and is composed of 34 distinct nucleoporin proteins. About half of the nucleoporins typically contain solenoid protein domains—either an alpha solenoid or a beta-propeller fold, or in some cases both as separate structural domains. The other half show structural characteristics typical of "natively unfolded" or intrinsically disordered proteins, i.e. they are highly flexible proteins that lack ordered tertiary structure. These disordered proteins are the FG nucleoporins, so called because their amino-acid sequence contains many phenylalanine–glycine repeats.
Okadaic acid, C44H68O13, is a toxin produced by several species of dinoflagellates, and is known to accumulate in both marine sponges and shellfish. One of the primary causes of diarrhetic shellfish poisoning, okadaic acid is a potent inhibitor of specific protein phosphatases and is known to have a variety of negative effects on cells. A polyketide, polyether derivative of a C38 fatty acid, okadaic acid and other members of its family have shined light upon many biological processes both with respect to dinoflagellete polyketide synthesis as well as the role of protein phosphatases in cell growth.

Hitachimycin, also known as stubomycin, is a cyclic polypeptide produced by Streptomyces that acts as an antibiotic. It exhibits cytotoxic activity against mammalian cells, Gram-positive bacteria, yeast, and fungi, as well as hemolytic activity; this is mediated by changes at the cell membrane and subsequent lysis. Owing to its cytotoxic activity against mammalian cells and tumors, it was first proposed as an antitumor antibiotic.
Leptomycins are secondary metabolites produced by Streptomyces spp.

Ran also known as GTP-binding nuclear protein Ran is a protein that in humans is encoded by the RAN gene. Ran is a small 25 kDa protein that is involved in transport into and out of the cell nucleus during interphase and also involved in mitosis. It is a member of the Ras superfamily.
Nuclear transport refers to the mechanisms by which molecules move across the nuclear membrane of a cell. The entry and exit of large molecules from the cell nucleus is tightly controlled by the nuclear pore complexes (NPCs). Although small molecules can enter the nucleus without regulation, macromolecules such as RNA and proteins require association with transport factors known as nuclear transport receptors, like karyopherins called importins to enter the nucleus and exportins to exit.

Rhizoxin is an antimitotic agent with anti-tumor activity. It is isolated from a pathogenic plant fungus which causes rice seedling blight.
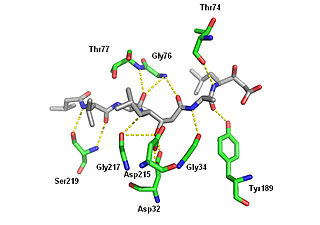
Pepstatin is a potent inhibitor of aspartyl proteases. It is a hexa-peptide containing the unusual amino acid statine, having the sequence Isovaleryl-Val-Val-Sta-Ala-Sta (Iva-Val-Val-Sta-Ala-Sta). It was originally isolated from cultures of various species of Actinomyces due to its ability to inhibit pepsin at picomolar concentrations. Pepstatin A is well known to be an inhibitor of aspartic proteases such as pepsin, cathepsins D and E. Except for its role as a protease inhibitor, however, the pharmacological action of pepstatin A upon cells remain unclear. Pepstatin A suppresses receptor activator of NF-κB ligand (RANKL)–induced osteoclast differentiation. Pepstatin A suppresses the formation of multinuclear osteoclasts dose-dependently. This inhibition of the formation only affected osteoclast cells, i.e., not osteoblast-like cells. Furthermore, pepstatin A also suppresses differentiation from pre-osteoclast cells to mononuclear osteoclast cells dose-dependently. This inhibition seems to be independent of the activities of proteases such as cathepsin D, because the formation of osteoclasts was not suppressed with the concentration that inhibited the activity of cathepsin D. Cell signaling analysis indicated that the phosphorylation of ERK was inhibited in pepstatin A-treated cells, while the phosphorylation of IκB and Akt showed almost no change. Furthermore, pepstatin A decreased the expression of nuclear factor of activated T cells c1 (NFATc1). These results suggest that pepstatin A suppresses the differentiation of osteoclasts through the blockade of ERK signaling and the inhibition of NFATc1 expression.
Neuroprotectin D1 (NPD1) also known as Protectin D1 (PD1) is a docosanoid derived from the polyunsaturated fatty acid (PUFA) docosahexaenoic acid (DHA), which is a component of fish oil and the most important omega-3 PUFA. Like other members of the specialized proresolving mediators class of PUFA metabolites, NPD1 exerts potent anti-inflammatory and anti-apoptotic/neuroprotective bioactivity. Other neuroprotectins with similar activity include: PDX ; 20-hydroxy-PD1 ; and 10-epi-PD1. The activity of neuroprotectin-like metabolite, 17-epi-PD1, has not yet been reported.

Exportin 1 (XPO1), also known as chromosomal region maintenance 1 (CRM1), is a eukaryotic protein that mediates the nuclear export of various proteins and RNAs.
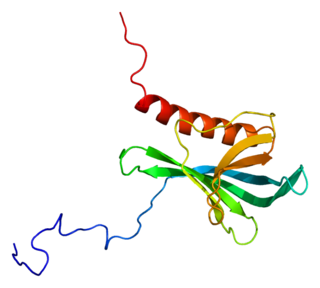
Ran-binding protein 3 is a protein that in humans is encoded by the RANBP3 gene.
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposite effect of a nuclear localization signal, which targets a protein located in the cytoplasm for import to the nucleus. The NES is recognized and bound by exportins.

Indolocarbazoles (ICZs) are a class of compounds that are under current study due to their potential as anti-cancer drugs and the prospective number of derivatives and uses found from the basic backbone alone. First isolated in 1977, a wide range of structures and derivatives have been found or developed throughout the world. Due to the extensive number of structures available, this review will focus on the more important groups here while covering their occurrence, biological activity, biosynthesis, and laboratory synthesis.

In organic chemistry, enediynes are organic compounds containing two triple bonds and one double bond.
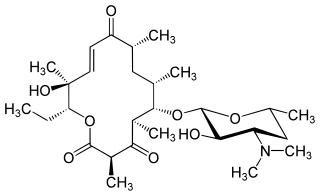
Pikromycin was studied by Brokmann and Hekel in 1951 and was the first antibiotic macrolide to be isolated. Pikromycin is synthesized through a type I polyketide synthase system in Streptomyces venezuelae, a species of Gram-positive bacterium in the genus Streptomyces. Pikromycin is derived from narbonolide, a 14-membered ring macrolide. Along with the narbonolide backbone, pikromycin includes a desosamine sugar and a hydroxyl group. Although Pikromycin is not a clinically useful antibiotic, it can be used as a raw material to synthesize antibiotic ketolide compounds such as ertythromycins and new epothilones.

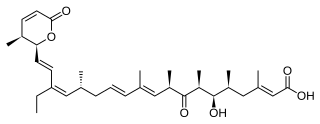
Callyspongia truncata is a species of marine sea sponge. Like all marine sponges, C. truncata is a member of phylum Porifera and is defined by its filter-feeding lifestyle and flagellated choanocytes, or collar cells, that allow for water movement and feeding. It is a species of demosponge and a member of Demospongiae, the largest class of sponges as well as the family Callyspongiidae. C. truncata is most well known for being the organism from which the polyketide Callystatin A was identified. Callystatin A is a polyketide natural product from the leptomycin family of antibiotics. It was first isolated in 1997 from this organism, which was collected from the Goto Islands in the Nagasaki Prefecture of Japan by the Kobayashi group. Recent studies have revealed numerous other bioactive compounds that have been found in this species.
Streptomyces cattleya is a Gram-positive bacterium which makes cephamycin, penicillin and thienamycin. The bacterium expresses a fluorinase enzyme, and the organism has been used to understand the biosynthesis of fluoroacetate and the antibacterial 4-fluoro-L-threonine. The γ-Glu-βes pathway to biosynthesis of non-traditional amino acids β-ethynylserine (βes) and L-propargylglycine (Pra) was first characterized in this species.

C-1027 or Lidamycin is an antitumor antibiotic consisting of a complex of an enediyne chromophore and an apoprotein. It shows antibiotic activity against most Gram-positive bacteria. It is one of the most potent cytotoxic molecules known, due to its induction of a higher ratio of DNA double-strand breaks than single-strand breaks.
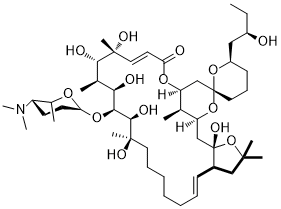
Ossamycin is a fermentation-derived natural product belonging to a family of 22- to 26-membered macrocyclic polyketides which is featured with a 6,6-spiroacetal (1,7-dioxaspiro[5,5]-undecanyl) moiety connected to one side of the macrocycle. Widely-studied 26-membered oligomycins/rutamycins, 24-membered dunaimycins, and 22-membered cytovaricin are also in this family.
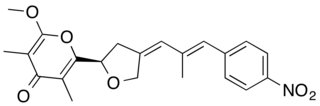
Aureothin is a natural product of a cytotoxic shikimate-polyketide antibiotic with the molecular formula C22H23NO6. Aureothin is produced by the bacterium Streptomyces thioluteus that illustrates antitumor, antifungal, and insecticidal activities and the new aureothin derivatives can be antifungal and antiproliferative. In addition, aureothin, a nitro compound from Streptomyces thioluteus, was indicated to have pesticidal activity against the bean weevil by interfering with mitochondrial respiratory complex II.




















