
Epoxide hydrolases (EHs), also known as epoxide hydratases, are enzymes that metabolize compounds that contain an epoxide residue; they convert this residue to two hydroxyl residues through an epoxide hydrolysis reaction to form diol products. Several enzymes possess EH activity. Microsomal epoxide hydrolase, soluble epoxide hydrolase, and the more recently discovered but not as yet well defined functionally, epoxide hydrolase 3 (EH3) and epoxide hydrolase 4 (EH4) are structurally closely related isozymes. Other enzymes with epoxide hydrolase activity include leukotriene A4 hydrolase, Cholesterol-5,6-oxide hydrolase, MEST (gene) (Peg1/MEST), and Hepoxilin-epoxide hydrolase. The hydrolases are distinguished from each other by their substrate preferences and, directly related to this, their functions.
The epoxyeicosatrienoic acids or EETs are signaling molecules formed within various types of cells by the metabolism of arachidonic acid by a specific subset of Cytochrome P450 enzymes termed cytochrome P450 epoxygenases. These nonclassic eicosanoids are generally short-lived, being rapidly converted from epoxides to less active or inactive dihydroxy-eicosatrienoic acids (diHETrEs) by a widely distributed cellular enzyme, Soluble epoxide hydrolase (sEH), also termed Epoxide hydrolase 2. The EETs consequently function as transiently acting, short-range hormones; that is, they work locally to regulate the function of the cells that produce them or of nearby cells. The EETs have been most studied in animal models where they show the ability to lower blood pressure possibly by a) stimulating arterial vasorelaxation and b) inhibiting the kidney's retention of salts and water to decrease intravascular blood volume. In these models, EETs prevent arterial occlusive diseases such as heart attacks and brain strokes not only by their anti-hypertension action but possibly also by their anti-inflammatory effects on blood vessels, their inhibition of platelet activation and thereby blood clotting, and/or their promotion of pro-fibrinolytic removal of blood clots. With respect to their effects on the heart, the EETs are often termed cardio-protective. Beyond these cardiovascular actions that may prevent various cardiovascular diseases, studies have implicated the EETs in the pathological growth of certain types of cancer and in the physiological and possibly pathological perception of neuropathic pain. While studies to date imply that the EETs, EET-forming epoxygenases, and EET-inactivating sEH can be manipulated to control a wide range of human diseases, clinical studies have yet to prove this. Determination of the role of the EETS in human diseases is made particularly difficult because of the large number of EET-forming epoxygenases, large number of epoxygenase substrates other than arachidonic acid, and the large number of activities, some of which may be pathological or injurious, that the EETs possess.
In enzymology, a cholestanetetraol 26-dehydrogenase (EC 1.1.1.161) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-keto-steroid reductase (EC 1.1.1.270) is an enzyme that catalyzes the chemical reaction
In enzymology, a cholestenone 5alpha-reductase (EC 1.3.1.22) is an enzyme that catalyzes the chemical reaction
In enzymology, a Delta14-sterol reductase (EC 1.3.1.70) is an enzyme that catalyzes the chemical reaction
In enzymology, a Delta24-sterol reductase (EC 1.3.1.72) is an enzyme that catalyzes the chemical reaction
In enzymology, a cholestanetriol 26-monooxygenase (EC 1.14.13.15) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3alpha,7alpha,12alpha-trihydroxycholestan-26-al 26-oxidoreductase is an enzyme that catalyzes the chemical reaction:
In enzymology, a cholestenol Δ-isomerase is an enzyme that catalyzes the chemical reaction
In enzymology, a hepoxilin-epoxide hydrolase is an enzyme that catalyzes the conversion of the epoxyalcohol metabolites arachidonic acid, hepoxilin A3 and hepoxilin B3 to their tri-hydroxyl products, trioxolin A3 and trioxilin B3, respectively. These reactions in general inactivate the two biologically active hepoxilins.

In enzymology, a microsomal epoxide hydrolase (mEH) is an enzyme that catalyzes the hydrolysis reaction between an epoxide and water to form a diol.

Cytochrome P450 4F8 is a protein that in humans is encoded by the CYP4F8 gene.

Epoxide hydrolase 1 is an enzyme encoded by the EPHX1 gene in humans.
Epoxygenases are a set of membrane-bound, heme-containing cytochrome P450 enzymes that metabolize polyunsaturated fatty acids to epoxide products that have a range of biological activities. The most thoroughly studied substrate of the CYP epoxylgenases is arachidonic acid. This polyunsaturated fatty acid is metabolized by cyclooxygenases to various prostaglandin, thromboxane, and prostacyclin metabolites in what has been termed the first pathway of eicosanoid production; it is also metabolized by various lipoxygenases to hydroxyeicosatetraenoic acids and leukotrienes in what has been termed the second pathway of eicosanoid production. The metabolism of arachidonic acid to epoxyeicosatrienoic acids by the CYP epoxygenases has been termed the third pathway of eicosanoid metabolism. Like the first two pathways of eicosanoid production, this third pathway acts as a signaling pathway wherein a set of enzymes metabolize arachidonic acid to a set of products that act as secondary signals to work in activating their parent or nearby cells and thereby orchestrate functional responses. However, none of these three pathways is limited to metabolizing arachidonic acid to eicosanoids. Rather, they also metabolize other polyunsaturated fatty acids to products that are structurally analogous to the eicosanoids but often have different bioactivity profiles. This is particularly true for the CYP epoxygenases which in general act on a broader range of polyunsaturated fatty acids to form a broader range of metabolites than the first and second pathways of eicosanoid production. Furthermore, the latter pathways form metabolites many of which act on cells by binding with and thereby activating specific and well-characterized receptor proteins; no such receptors have been fully characterized for the epoxide metabolites. Finally, there are relatively few metabolite-forming lipoxygenases and cyclooxygenases in the first and second pathways and these oxygenase enzymes share similarity between humans and other mammalian animal models. The third pathway consists of a large number of metabolite-forming CYP epoxygenases and the human epoxygenases have important differences from those of animal models. Partly because of these differences, it has been difficult to define clear roles for the epoxygenase-epoxide pathways in human physiology and pathology.

Soluble epoxide hydrolase (sEH) is a bifunctional enzyme that in humans is encoded by the EPHX2 gene. sEH is a member of the epoxide hydrolase family. This enzyme, found in both the cytosol and peroxisomes, binds to specific epoxides and converts them to the corresponding diols. A different region of this protein also has lipid-phosphate phosphatase activity. Mutations in the EPHX2 gene have been associated with familial hypercholesterolemia.
Methylsterol monooxygenase (EC 1.14.13.72, methylsterol hydroxylase, 4-methylsterol oxidase, 4,4-dimethyl-5alpha-cholest-7-en-3beta-ol,hydrogen-donor:oxygen oxidoreductase (hydroxylating)) is an enzyme with systematic name 4,4-dimethyl-5alpha-cholest-7-en-3beta-ol,NAD(P)H:oxygen oxidoreductase (hydroxylating). This enzyme catalyses the following chemical reaction
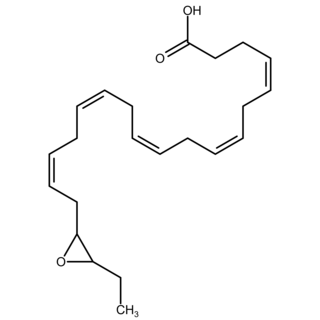
Epoxide docosapentaenoic acids are metabolites of the 22-carbon straight-chain omega-3 fatty acid, docosahexaenoic acid (DHA). Cell types that express certain cytochrome P450 (CYP) epoxygenases metabolize polyunsaturated fatty acid's (PUFAs) by converting one of their double bonds to an epoxide. In the best known of these metabolic pathways, cellular CYP epoxygenases metabolize the 20-carbon straight-chain omega-6 fatty acid, arachidonic acid, to epoxyeicosatrienoic acids (EETs); another CYP epoxygenase pathway metabolizes the 20-carbon omega-3 fatty acid, eicosapentaenoic acid (EPA), to epoxyeicosatetraenoic acids (EEQs). CYP epoxygenases similarly convert various other PUFAs to epoxides These epoxide metabolites have a variety of activities. However, essentially all of them are rapidly converted to their corresponding, but in general far less active, Vicinal (chemistry) dihydroxy fatty acids by ubiquitous cellular Soluble epoxide hydrolase. Consequently, these epoxides, including EDPs, operate as short-lived signaling agents that regulate the function of their parent or nearby cells. The particular feature of EDPs distinguishing them from EETs is that they derive from omega-3 fatty acids and are suggested to be responsible for some of the beneficial effects attributed to omega-3 fatty acids and omega-3-rich foods such as fish oil.

Epoxyeicosatetraenoic acids are a set of biologically active epoxides that various cell types make by metabolizing the omega 3 fatty acid, eicosapentaenoic acid (EPA), with certain cytochrome P450 epoxygenases. These epoxygenases can metabolize EPA to as many as 10 epoxides that differ in the site and/or stereoisomer of the epoxide formed; however, the formed EEQs, while differing in potency, often have similar bioactivities and are commonly considered together.

Epoxide hydrolase 3 is a protein that in humans is encoded by the EPHX3 gene. It is the third defined isozyme in a set of epoxide hydrolase isozymes, the epoxide hydrolases. This set includes the Microsomal epoxide hydrolase ; the epoxide hydrolase 2 ; and the far less well defined enzymatically, epoxide hydrolase 4. All four enzyme contain an Alpha/beta hydrolase fold suggesting that they have Hydrolysis activity. EH1, EH2, and EH3 have been shown to have such activity in that they add water to epoxides of unsaturated fatty acids to form vicinal cis products; the activity of EH4 has not been reported. The former three EH's differ in subcellular location, tissue expression patterns, substrate preferences, and thereby functions. These functions include limiting the biologically actions of certain fatty acid epoxides, increasing the toxicity of other fatty acid epoxides, and contributing to the metabolism of drugs and other xenobiotics.







