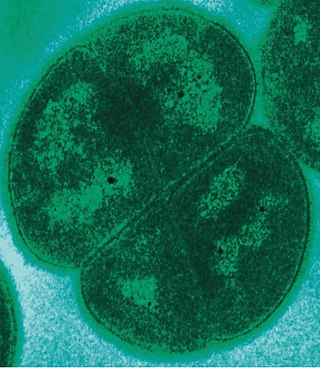
Deinococcota is a phylum of bacteria with a single class, Deinococci, that are highly resistant to environmental hazards, also known as extremophiles. These bacteria have thick cell walls that give them gram-positive stains, but they include a second membrane and so are closer in structure to those of gram-negative bacteria.

Desiccation is the state of extreme dryness, or the process of extreme drying. A desiccant is a hygroscopic substance that induces or sustains such a state in its local vicinity in a moderately sealed container. The word desiccation comes from Latin de- 'thoroughly', and siccare 'to dry'.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encodes its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.
Radioresistance is the level of ionizing radiation that organisms are able to withstand.
Radiosensitivity is the relative susceptibility of cells, tissues, organs or organisms to the harmful effect of ionizing radiation.

Halobacterium is a genus in the family Halobacteriaceae.
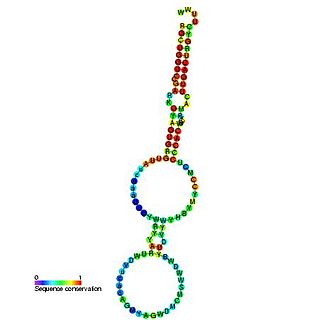
Y RNAs are small non-coding RNAs. They are components of the Ro60 ribonucleoprotein particle which is a target of autoimmune antibodies in patients with systemic lupus erythematosus. They are also reported to be necessary for DNA replication through interactions with chromatin and initiation proteins. However, mouse embryonic stem cells lacking Y RNAs are viable and have normal cell cycles.

Evelyn M. Witkin was an American bacterial geneticist at Cold Spring Harbor Laboratory (1944–1955), SUNY Downstate Medical Center (1955–1971), and Rutgers University (1971–1991). Witkin was considered innovative and inspirational as a scientist, teacher and mentor.
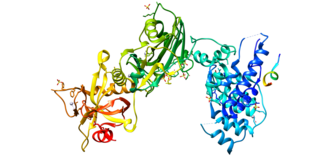
Artemis is a protein that in humans is encoded by the DCLRE1C gene.

Deinococcus radiodurans is a bacterium, an extremophile and one of the most radiation-resistant organisms known. It can survive cold, dehydration, vacuum, and acid, and therefore is known as a polyextremophile. The Guinness Book Of World Records listed it as the world's toughest known bacterium.

Deinococcus is in the monotypic family Deinococcaceae, and one genus of three in the order Deinococcales of the bacterial phylum Deinococcota highly resistant to environmental hazards. These bacteria have thick cell walls that give them Gram-positive stains, but they include a second membrane and so are closer in structure to Gram-negative bacteria. Deinococcus survive when their DNA is exposed to high doses of gamma and UV radiation. Whereas other bacteria change their structure in the presence of radiation, such as by forming endospores, Deinococcus tolerate it without changing their cellular form and do not retreat into a hardened structure. They are also characterized by the presence of the carotenoid pigment deinoxanthin that give them their pink color. They are usually isolated according to these two criteria. In August 2020, scientists reported that bacteria from Earth, particularly Deinococcus bacteria, were found to survive for three years in outer space, based on studies conducted on the International Space Station. These findings support the notion of panspermia, the hypothesis that life exists throughout the Universe, distributed in various ways, including space dust, meteoroids, asteroids, comets, planetoids or contaminated spacecraft.

Chroococcidiopsis is a photosynthetic, coccoidal bacterium, and the only genus in the order Chroococcidiopsidales and in the family Chroococcidiopsidaceae. A diversity of species and cultures exist within the genus, with a diversity of phenotypes. Some extremophile members of Chroococcidiopsis are known for their ability to survive harsh environmental conditions, including both high and low temperatures, ionizing radiation, and high salinity.
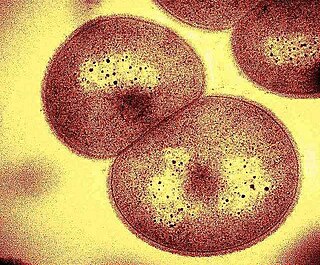
Deinococcus geothermalis is a non-pathogenic, sphere-shaped, Gram-positive, heterotrophic bacterium, where geothermalis means 'hot earth' or 'hot springs'. This bacterium was first obtained from the hot springs of Agnano, Naples, Italy and São Pedro do Sul, Portugal. It resides primarily in hot springs and in deep ocean environments.
Deinococcus ficus strain CC-FR2-10T is a recently discovered gram-positive bacteria which plays a role in the production of nitrogen fertilizer. It was originally isolated from a Ficus plant, hence its name.
BASys is a freely available web server that can be used to perform automated, comprehensive annotation of bacterial genomes. With the advent of next generation DNA sequencing it is now possible to sequence the complete genome of a bacterium within a single day. This has led to an explosion in the number of fully sequenced microbes. In fact, as of 2013, there were more than 2700 fully sequenced bacterial genomes deposited with GenBank. However, a continuing challenge with microbial genomics is finding the resources or tools for annotating the large number of newly sequenced genomes. BASys was developed in 2005 in anticipation of these needs. In fact, BASys was the world’s first publicly accessible microbial genome annotation web server. Because of its widespread popularity, the BASys server was updated in 2011 through the addition of multiple server nodes to handle the large number of queries it was receiving.
Deinococcus frigens is a species of low temperature and drought-tolerating, UV-resistant bacteria from Antarctica. It is Gram-positive, non-motile and coccoid-shaped. Its type strain is AA-692. Individual Deinococcus frigens range in size from 0.9-2.0 μm and colonies appear orange or pink in color. Liquid-grown cells viewed using phase-contrast light microscopy and transmission electron microscopy on agar-coated slides show that isolated D. frigens appear to produce buds. Comparison of the genomes of Deiococcus radiodurans and D. frigens have predicted that no flagellar assembly exists in D. frigens.
Deinococcus marmoris is a Gram-positive bacterium isolated from Antarctica. As a species of the genus Deinococcus, the bacterium is UV-tolerant and able to withstand low temperatures.
Halorhodospira halophila is a species of Halorhodospira distinguished by its ability to grow optimally in an environment of 15–20% salinity. It was formerly called Ectothiorhodospira halophila. It is an anaerobic, rod-shaped Gram-negative bacterium. H. halophila has a flagellum.
Ionizing radiation can cause biological effects which are passed on to offspring through the epigenome. The effects of radiation on cells has been found to be dependent on the dosage of the radiation, the location of the cell in regards to tissue, and whether the cell is a somatic or germ line cell. Generally, ionizing radiation appears to reduce methylation of DNA in cells.
Deinococcus aerius is an anaerobic bacterium that can be found in the atmosphere above the island of Japan. Living in such conditions makes these bacteria highly resistant to desiccation, UV-C, and gamma radiation. Although previously unidentified as strain TR0125, this bacterium was determined to be Deinococcus aerius by 16S rRNA sequencing.








