Related Research Articles

Agarose gel electrophoresis is a method of gel electrophoresis used in biochemistry, molecular biology, genetics, and clinical chemistry to separate a mixed population of macromolecules such as DNA or proteins in a matrix of agarose, one of the two main components of agar. The proteins may be separated by charge and/or size, and the DNA and RNA fragments by length. Biomolecules are separated by applying an electric field to move the charged molecules through an agarose matrix, and the biomolecules are separated by size in the agarose gel matrix.

Gel electrophoresis is a method for separation and analysis of biomacromolecules and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.
Polyacrylamide gel electrophoresis (PAGE) is a technique widely used in biochemistry, forensic chemistry, genetics, molecular biology and biotechnology to separate biological macromolecules, usually proteins or nucleic acids, according to their electrophoretic mobility. Electrophoretic mobility is a function of the length, conformation, and charge of the molecule. Polyacrylamide gel electrophoresis is a powerful tool used to analyze RNA samples. When polyacrylamide gel is denatured after electrophoresis, it provides information on the sample composition of the RNA species.

The western blot, or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detecting the proteins, this technique is also utilized to visualize, distinguish, and quantify the different proteins in a complicated protein combination.

Gel electrophoresis of nucleic acids is an analytical technique to separate DNA or RNA fragments by size and reactivity. Nucleic acid molecules are placed on a gel, where an electric field induces the nucleic acids to migrate toward the positively charged anode. The molecules separate as they travel through the gel based on the each molecule's size and shape. Longer molecules move more slowly because they the gel resists their movement more forcefully than it resists shorter molecules. After some time, the electricity is turned off and the positions of the different molecules are analyzed.

A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles of a specific DNA sequence, known as probes. These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA sample under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis. It is also used for the identification of structural variations and the measurement of gene expression.

Two-dimensional gel electrophoresis, abbreviated as 2-DE or 2-D electrophoresis, is a form of gel electrophoresis commonly used to analyze proteins. Mixtures of proteins are separated by two properties in two dimensions on 2D gels. 2-DE was first independently introduced by O'Farrell and Klose in 1975.

Isoelectric focusing (IEF), also known as electrofocusing, is a technique for separating different molecules by differences in their isoelectric point (pI). It is a type of zone electrophoresis usually performed on proteins in a gel that takes advantage of the fact that overall charge on the molecule of interest is a function of the pH of its surroundings.

Immunoelectrophoresis is a general name for a number of biochemical methods for separation and characterization of proteins based on electrophoresis and reaction with antibodies. All variants of immunoelectrophoresis require immunoglobulins, also known as antibodies, reacting with the proteins to be separated or characterized. The methods were developed and used extensively during the second half of the 20th century. In somewhat chronological order: Immunoelectrophoretic analysis, crossed immunoelectrophoresis, rocket-immunoelectrophoresis, fused rocket immunoelectrophoresis ad modum Svendsen and Harboe, affinity immunoelectrophoresis ad modum Bøg-Hansen.
Protein methods are the techniques used to study proteins. There are experimental methods for studying proteins. Computational methods typically use computer programs to analyze proteins. However, many experimental methods require computational analysis of the raw data.
Cyanines, also referred to as tetramethylindo(di)-carbocyanines are a synthetic dye family belonging to the polymethine group. Although the name derives etymologically from terms for shades of blue, the cyanine family covers the electromagnetic spectrum from near IR to UV.

A molecular-weight size marker, also referred to as a protein ladder, DNA ladder, or RNA ladder, is a set of standards that are used to identify the approximate size of a molecule run on a gel during electrophoresis, using the principle that molecular weight is inversely proportional to migration rate through a gel matrix. Therefore, when used in gel electrophoresis, markers effectively provide a logarithmic scale by which to estimate the size of the other fragments.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.
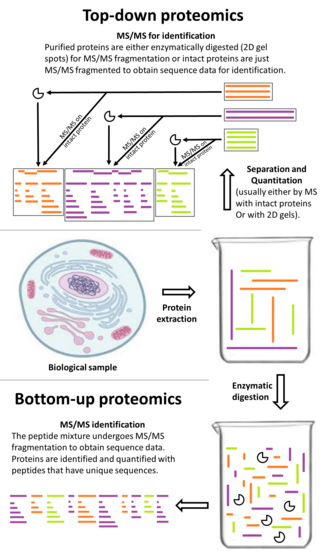
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE), preparative native PAGE, or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.

An electrophoretic color marker is a chemical used to monitor the progress of agarose gel electrophoresis and polyacrylamide gel electrophoresis (PAGE) since DNA, RNA, and most proteins are colourless. The color markers are made up of a mixture of dyes that migrate through the gel matrix alongside the sample of interest. They are typically designed to have different mobilities from the sample components and to generate colored bands that can be used to assess the migration and separation of sample components.
Free-flow electrophoresis (FFE), also known as carrier-free electrophoresis, is a matrix-free, high-voltage electrophoretic separation technique. FFE is an analogous technique to capillary electrophoresis, with a comparable resolution, that can be used for scientific questions, where semi-preparative and preparative amounts of samples are needed. It is used to quantitatively separate samples according to differences in charge or isoelectric point by forming a pH gradient. Because of the versatility of the technique, a wide range of protocols for the separation of samples like rare metal ions, protein isoforms, multiprotein complexes, peptides, organelles, cells, DNA origami, blood serum and nanoparticles exist. The advantage of FFE is the fast and gentle separation of samples dissolved in a liquid solvent without any need of a matrix, like polyacrylamide in gel electrophoresis. This ensures a very high recovery rate since analytes do not adhere to any carrier or matrix structure. Because of its continuous nature and high volume throughput, this technique allows a fast separation of preparative amounts of samples with a very high resolution. Furthermore, the separations can be conducted under native or denaturing conditions.
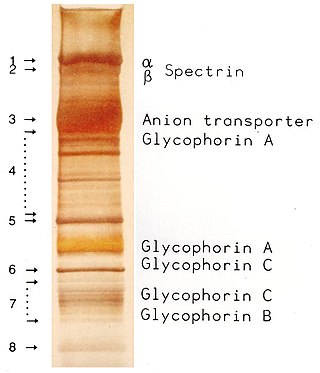
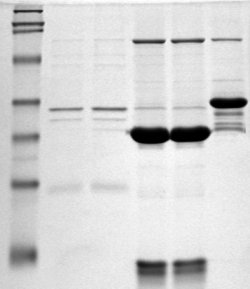
SDS-PAGE is a discontinuous electrophoretic system developed by Ulrich K. Laemmli which is commonly used as a method to separate proteins with molecular masses between 5 and 250 kDa. The combined use of sodium dodecyl sulfate and polyacrylamide gel eliminates the influence of structure and charge, and proteins are separated by differences in their size. At least up to 2012, the publication describing it was the most frequently cited paper by a single author, and the second most cited overall.
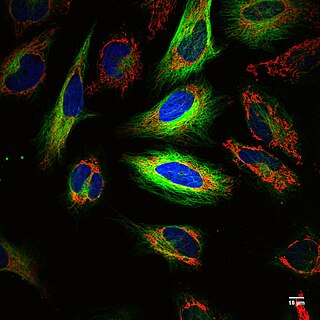
Fluorescence imaging is a type of non-invasive imaging technique that can help visualize biological processes taking place in a living organism. Images can be produced from a variety of methods including: microscopy, imaging probes, and spectroscopy.
References
- ↑ Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997 Oct;18(11):2071-7. PMID 9420172
- ↑ Alban, Andrew; David, Stephen Olu; Bjorkesten, Lennart; Andersson, Christian; Sloge, Erik; Lewis, Steve; Currie, Ian (2003). "A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard". Proteomics. 3 (1): 36–44. doi:10.1002/pmic.200390006. ISSN 1615-9853. PMID 12548632.