
Ricin ( RY-sin) is a lectin (a carbohydrate-binding protein) and a highly potent toxin produced in the seeds of the castor oil plant, Ricinus communis. The median lethal dose (LD50) of ricin for mice is around 22 micrograms per kilogram of body weight via intraperitoneal injection. Oral exposure to ricin is far less toxic. An estimated lethal oral dose in humans is approximately one milligram per kilogram of body weight.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Transfer-messenger RNA is a bacterial RNA molecule with dual tRNA-like and messenger RNA-like properties. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with Small Protein B (SmpB), Elongation Factor Tu (EF-Tu), and ribosomal protein S1. In trans-translation, tmRNA and its associated proteins bind to bacterial ribosomes which have stalled in the middle of protein biosynthesis, for example when reaching the end of a messenger RNA which has lost its stop codon. The tmRNA is remarkably versatile: it recycles the stalled ribosome, adds a proteolysis-inducing tag to the unfinished polypeptide, and facilitates the degradation of the aberrant messenger RNA. In the majority of bacteria these functions are carried out by standard one-piece tmRNAs. In other bacterial species, a permuted ssrA gene produces a two-piece tmRNA in which two separate RNA chains are joined by base-pairing.

Abrin is an extremely toxic toxalbumin found in the seeds of the rosary pea, Abrus precatorius. It has a median lethal dose of 0.7 micrograms per kilogram of body mass when given to mice intravenously. The median toxic dose for humans ranges from 10 to 1000 micrograms per kilogram when ingested and is 3.3 micrograms per kilogram when inhaled.

Angiogenin (ANG) also known as ribonuclease 5 is a small 123 amino acid protein that in humans is encoded by the ANG gene. Angiogenin is a potent stimulator of new blood vessels through the process of angiogenesis. Ang hydrolyzes cellular RNA, resulting in modulated levels of protein synthesis and interacts with DNA causing a promoter-like increase in the expression of rRNA. Ang is associated with cancer and neurological disease through angiogenesis and through activating gene expression that suppresses apoptosis.
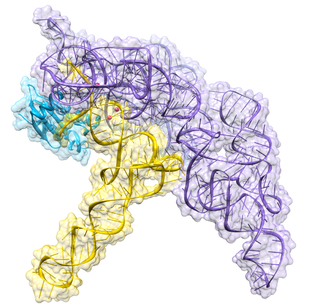
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.

Ribonuclease T1 (EC 4.6.1.24, guanyloribonuclease, Aspergillus oryzae ribonuclease, RNase N1, RNase N2, ribonuclease N3, ribonuclease U1, ribonuclease F1, ribonuclease Ch, ribonuclease PP1, ribonuclease SA, RNase F1, ribonuclease C2, binase, RNase Sa, guanyl-specific RNase, RNase G, RNase T1, ribonuclease guaninenucleotido-2'-transferase (cyclizing), ribonuclease N3, ribonuclease N1) is a fungal endonuclease that cleaves single-stranded RNA after guanine residues, i.e., on their 3' end; the most commonly studied form of this enzyme is the version found in the mold Aspergillus oryzae. Owing to its specificity for guanine, RNase T1 is often used to digest denatured RNA prior to sequencing. Similar to other ribonucleases such as barnase and RNase A, ribonuclease T1 has been popular for folding studies.

The 5S ribosomal RNA is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life, with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S).
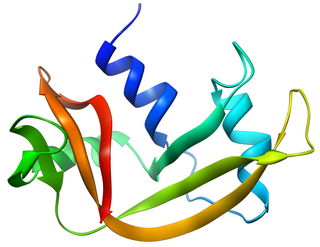
Pancreatic ribonuclease family is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles.
Saporin is a protein that is useful in biological research applications, especially studies of behavior. Saporins are so-called ribosome inactivating proteins (RIPs), due to its N-glycosidase activity, from the seeds of Saponaria officinalis. It was first described by Fiorenzo Stirpe and his colleagues in 1983 in an article that illustrated the unusual stability of the protein.

EF-G is a prokaryotic elongation factor involved in mRNA translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.
Beetin is a ribosome-inactivating protein found in the leaves of sugar beets, Beta vulgaris L, specifically attacking plant ribosomes. Sugar beet, beetins, that have been isolated meet all the criteria to be classified as single chain ribosome inactivating proteins that are highly toxic to mammalian ribosomes but non-toxic to intact cultured mammalian cells. Beetin expression occurs when there is a viral infection of the plant. The different levels of glycosylation of the same polypeptide chain result in the two forms of beetin. Beetin exhibits these two primary forms with apparent Mr values of 27 000 (BE27) and 29 000 (BE29) along with possessing glycan chains. Beetins are a type-I (single-chain) proteins with N-glycoside activity. Since it has been discovered that beetin is mostly concentrated in the intercellular fluid, its presence in the remaining parts of the leaf may be below the limit of detection rather than being nonexistent. The expression of beetin is only found in mature plants, but is present in all developing stages.

A ribosome-inactivating protein (RIP) is a protein synthesis inhibitor that acts at the eukaryotic ribosome. This protein family describes a large family of such proteins that work by acting as rRNA N-glycosylase. They inactivate 60S ribosomal subunits by an N-glycosidic cleavage, which releases a specific adenine base from the sugar-phosphate backbone of 28S rRNA. RIPs exist in bacteria and plants.

The antifungal proteinfamily is a protein family, with members sharing a structure consisting of five antiparallel beta strands which are highly twisted creating a beta barrel stabilised by four internal disulphide bridges. A cationic site adjacent to a hydrophobic stretch on the protein surface may constitute a phospholipid binding site.
rRNA endonuclease is an enzyme that catalyses the hydrolysis of the phosphodiester linkage between guanosine and adenosine residues at one specific position in the 28S rRNA of rat ribosomes. This enzyme also acts on bacterial rRNA.
Internal-loops in RNA are found where the double stranded RNA separates due to no Watson-Crick-Franklin base pairing between the nucleotides. Internal-loops differ from Stem-loops as they occur in middle of a stretch of double stranded RNA. The non-canonicoal residues result in the double helix becoming distorted due to unwinding, unstacking and kinking.
The SraL RNA, also known as RyjA, is a small non-coding RNA discovered in E. coli, and later in Salmonella Tiphimurium. This ncRNA was found to be expressed only in stationary phase. It may possibly play a role in Salmonella virulence. The major stationary phase regulator RpoS is transcriptionally regulating SraL and directly binds to the sraL gene promoter. SraL down-regulates the expression of the ribosome-associated chaperone Trigger Factor (TF), which is involved in the folding of the newly synthesised cystolic proteins.
Aspergillus giganteus is a species of fungus in the genus Aspergillus that grows as a mold. It was first described in 1901 by Wehmer, and is one of six Aspergillus species from the Clavati section of the subgenus Fumigati. Its closest taxonomic relatives are Aspergillus rhizopodus and Aspergillus longivescia.













